Predicting appropriateness of antibiotic treatment among ICU patients with hospital-acquired infection
- PMID: 39915601
- PMCID: PMC11802796
- DOI: 10.1038/s41746-024-01426-9
Predicting appropriateness of antibiotic treatment among ICU patients with hospital-acquired infection
Abstract
Antimicrobial resistance is a rising global health threat, leading to ineffective treatments, increased mortality and rising healthcare costs. In ICUs, inappropriate empiric antibiotic therapy is often given due to treatment urgency, causing poor outcomes. This study developed a machine learning model to predict the appropriateness of empiric antibiotics for ICU-acquired bloodstream infections, using data from the MIMIC-III database. To address missing values and dataset imbalances, novel computational methods were introduced. The model achieved an AUROC of 77.3% and AUPRC of 40.4% on validation, with similar results on external datasets from MIMIC-IV and Rambam Hospital. The model also predicted mortality risk, identifying a 30% mortality rate in high-risk patients versus 16.8% in low-risk groups. External validation on the eICU database showed a comparable gap, with mortality rates at 24% for high-risk and 7.7% for low-risk groups. Our study demonstrates the potential of machine learning models to predict inappropriate empiric antibiotic treatment.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
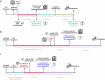
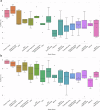
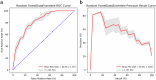
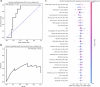
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

