Targeted detection of endogenous LINE-1 proteins and ORF2p interactions
- PMID: 39915890
- PMCID: PMC11800616
- DOI: 10.1186/s13100-024-00339-4
Targeted detection of endogenous LINE-1 proteins and ORF2p interactions
Abstract
Background: Both the expression and activities of LINE-1 (L1) retrotransposons are known to occur in numerous cell-types and are implicated in pathobiological contexts such as aging-related inflammation, autoimmunity, and in cancers. L1s encode two proteins that are translated from bicistronic transcripts. The translation product of ORF1 (ORF1p) has been robustly detected by immunoassays and shotgun mass spectrometry (MS). Yet, more sensitive detection methods would enhance the use of ORF1p as a clinical biomarker. In contrast, until now, no direct evidence of endogenous L1 ORF2 translation to protein (ORF2p) has been shown. Instead, assays for ORF2p have been limited to ectopic L1 ORF over-expression contexts and to indirect detection of endogenous ORF2p enzymatic activity, such as by the sequencing of de novo genomic insertions. Immunoassays for endogenous ORF2p have been problematic, producing apparent false positives due to cross-reactivities, and shotgun MS has not yielded reliable evidence of ORF2p peptides in biological samples.
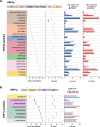
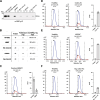
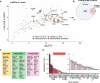
Results: Here we present targeted mass spectrometry assays, selected and parallel reaction monitoring (SRM and PRM, respectively) to detect and quantify L1 ORF1p and ORF2p at their endogenous abundances. We were able to quantify ORF1p and ORF2p present in our samples down to a range in the low attomoles. Confident in our ability to affinity enrich ORF2p, we describe an interactome associated with endogenous ORF2-containing macromolecular assemblies.
Conclusions: This is the first assay to demonstrate sensitive and robust quantitation of endogenous ORF2p. The ability to assay ORF2p directly and quantitatively will improve our understanding of the developmental and diseased cell states where L1 expression and its activity naturally occur. The ability to simultaneously assay endogenous L1 ORF1p and ORF2p is an important step forward for L1 analytical biochemistry. Endogenous ORF2p interactomes can now be presented with confidence that ORF2p is among the enriched proteins.
Keywords: Cancer; LINE-1; Mass spectrometry; PRM; Retrotransposon; SRM; Targeted proteomics.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Fresh frozen tissues were collected at Massachusetts General Hospital Department of Pathology as de-identified patient samples in accordance with Exemption 4, of research involving human subjects, from the National Institutes of Health. These samples were subsequently analyzed at The Rockefeller University, where, according to 45 CFR 46.102 (f) of the U.S. Dept. of Health and Human Services, it was determined that this research does not involve human subjects (IRB reference #334332). Consent for publication: Not applicable. Competing interests: JL reports grants, personal fees, and equity from Rome Therapeutics, outside the submitted work. MT reports personal fees, and equity from Rome Therapeutics, outside the submitted work. JL, MIN, and JCW have a patent application pending, based on this work. The other authors declare no competing interests.
Figures


Similar articles
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
[Volume and health outcomes: evidence from systematic reviews and from evaluation of Italian hospital data].Epidemiol Prev. 2013 Mar-Jun;37(2-3 Suppl 2):1-100. Epidemiol Prev. 2013. PMID: 23851286 Italian.
-
Incentives for preventing smoking in children and adolescents.Cochrane Database Syst Rev. 2017 Jun 6;6(6):CD008645. doi: 10.1002/14651858.CD008645.pub3. Cochrane Database Syst Rev. 2017. PMID: 28585288 Free PMC article.
References
-
- Feng Q, Moran JV, Kazazian HH, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–16. - PubMed
-
- Mathias SL, Scott AF, Kazazian HH, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–10. - PubMed
-
- Ostertag EM, Kazazian HH. Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–38. - PubMed
-
- Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

