Transgenerational epigenetic heritability for growth, body composition, and reproductive traits in Landrace pigs
- PMID: 39917178
- PMCID: PMC11799271
- DOI: 10.3389/fgene.2024.1526473
Transgenerational epigenetic heritability for growth, body composition, and reproductive traits in Landrace pigs
Abstract
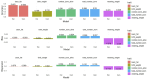
Epigenetics is an important source of variation in complex traits that is not due to changes in DNA sequences, and is dependent on the environment the individuals are exposed to. Therefore, we aimed to estimate transgenerational epigenetic heritability, percentage of resetting epigenetic marks, genetic parameters, and predicting breeding values using genetic and epigenetic models for growth, body composition, and reproductive traits in Landrace pigs using routinely recorded datasets. Birth and weaning weight, backfat thickness, total number of piglets born, and number of piglets born alive (BW, WW, BF, TNB, and NBA, respectively) were investigated. Models including epigenetic effects had a similar or better fit than solely genetic models. Including genomic information in epigenetic models resulted in large changes in the variance component estimates. Transgenerational epigenetic heritability estimates ranged between 0.042 (NBA) to 0.336 (BF). The reset coefficient estimates for epigenetic marks were between 80% and 90%. Heritability estimates for the direct additive and maternal genetic effects ranged between 0.040 (BW) to 0.502 (BF) and 0.034 (BF) to 0.134 (BW), respectively. Repeatability of the reproductive traits ranged between 0.098 (NBA) to 0.148 (TNB). Prediction accuracies, bias, and dispersion of breeding values ranged between 0.199 (BW) to 0.443 (BF), -0.080 (WW) to 0.034 (NBA), and -0.134 (WW) to 0.131 (TNB), respectively, with no substantial differences between genetic and epigenetic models. Transgenerational epigenetic heritability estimates are moderate for growth and body composition and low for reproductive traits in North American Landrace pigs. Fitting epigenetic effects in genetic models did not impact the prediction of breeding values.
Keywords: ebv; epigenetics; genetic parameters; genomics; swine; variance components.
Copyright © 2025 Araujo, Johnson, Graham, Howard, Huang, Oliveira and Brito.
Conflict of interest statement
Authors JH and YH were employed by the company Smithfield Premium Genetics. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures



References
-
- Akaike H. (1974). A new look at the statistical model identification. IEEE Trans. Autom. Control 19, 716–723. 10.1109/TAC.1974.1100705 - DOI
-
- Akanno E. C., Schenkel F. S., Quinton V. M., Friendship R. M., Robinson J. A. B. (2013). Meta-analysis of genetic parameter estimates for reproduction, growth and carcass traits of pigs in the tropics. Livest. Sci. 152, 101–113. 10.1016/j.livsci.2012.07.021 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

