Impact of developmental state, p53 status, and interferon signaling on glioblastoma cell response to radiation and temozolomide treatment
- PMID: 39919036
- PMCID: PMC11805374
- DOI: 10.1371/journal.pone.0315171
Impact of developmental state, p53 status, and interferon signaling on glioblastoma cell response to radiation and temozolomide treatment
Abstract
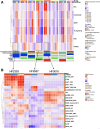
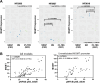
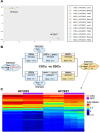
Glioblastoma (GBM) tumors exhibit extensive genomic, epigenomic, and transcriptional diversity, with significant intratumoral heterogeneity, complicating standard treatment approaches involving radiation (RT) and the DNA-alkylating agent temozolomide (TMZ). In this study, we employed an integrative multi-omics approach, including targeted proteomics, transcriptomics, genomics, and DNA methylation profiling, to investigate the response of a representative panel of GBM patient-derived cancer stem cells (CSCs) to astrocytic differentiation and RT and TMZ treatments. Differentiated CSC progenies retained the expression of key stemness genes and survival pathways, while activating the BMP-Smad signaling pathway and upregulating extracellular matrix components. This was associated with increased resistance to TMZ, though not to RT, across all models. We identified TP53 status as a critical determinant of transcriptional response to both RT and TMZ, which was also modulated by the differentiation state and treatment modality in wildtype (wt) p53 GBM cells. Both mutant and wt p53 models exhibited significant activation of the DNA-damage associated interferon (IFN) response in CSCs and differentiated cells, implicating this pathway in the GBM response to therapy. We observed that activation of NF-κB was positively correlated with the levels of O-6-methylguanine-DNA methyltransferase (MGMT) protein, a direct DNA repair enzyme leading to TMZ resistance, regardless of MGMT promoter methylation status, further supporting the clinical potential for inhibition of NF-kB signaling in GBM treatment. Our integrative analysis of the impact of GBM cell developmental states, in the context of genomic and molecular diversity of patient-derived models, provides valuable insights for pre-clinical studies aimed at optimizing treatment strategies.
Copyright: © 2025 Berezovsky et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Marko NF, Weil RJ, Schroeder JL, Lang FF, Suki D, Sawaya RE. Extent of resection of glioblastoma revisited: personalized survival modeling facilitates more accurate survival prediction and supports a maximum-safe-resection approach to surgery. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2014;32(8):774–82. Epub 20140210. doi: 10.1200/JCO.2013.51.8886 ; PubMed Central PMCID: PMC4876349. - DOI - PMC - PubMed
-
- Marenco-Hillembrand L, Wijesekera O, Suarez-Meade P, Mampre D, Jackson C, Peterson J, et al.. Trends in glioblastoma: outcomes over time and type of intervention: a systematic evidence based analysis. Journal of neuro-oncology. 2020;147(2):297–307. Epub 2020/03/12. doi: 10.1007/s11060-020-03451-6 . - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

