Constant pH Simulation with FMM Electrostatics in GROMACS. (A) Design and Applications
- PMID: 39919102
- PMCID: PMC11866755
- DOI: 10.1021/acs.jctc.4c01318
Constant pH Simulation with FMM Electrostatics in GROMACS. (A) Design and Applications
Abstract
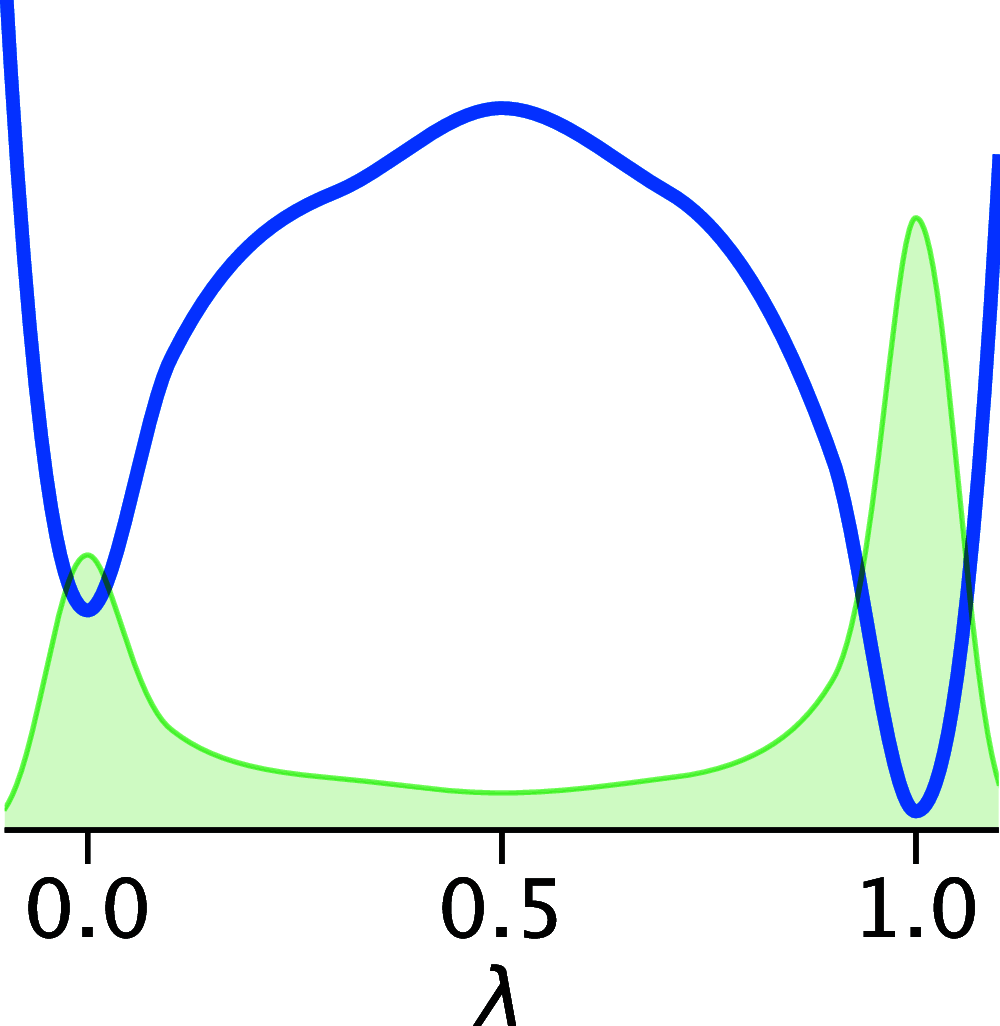
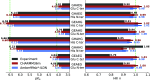
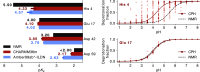
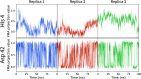
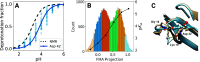
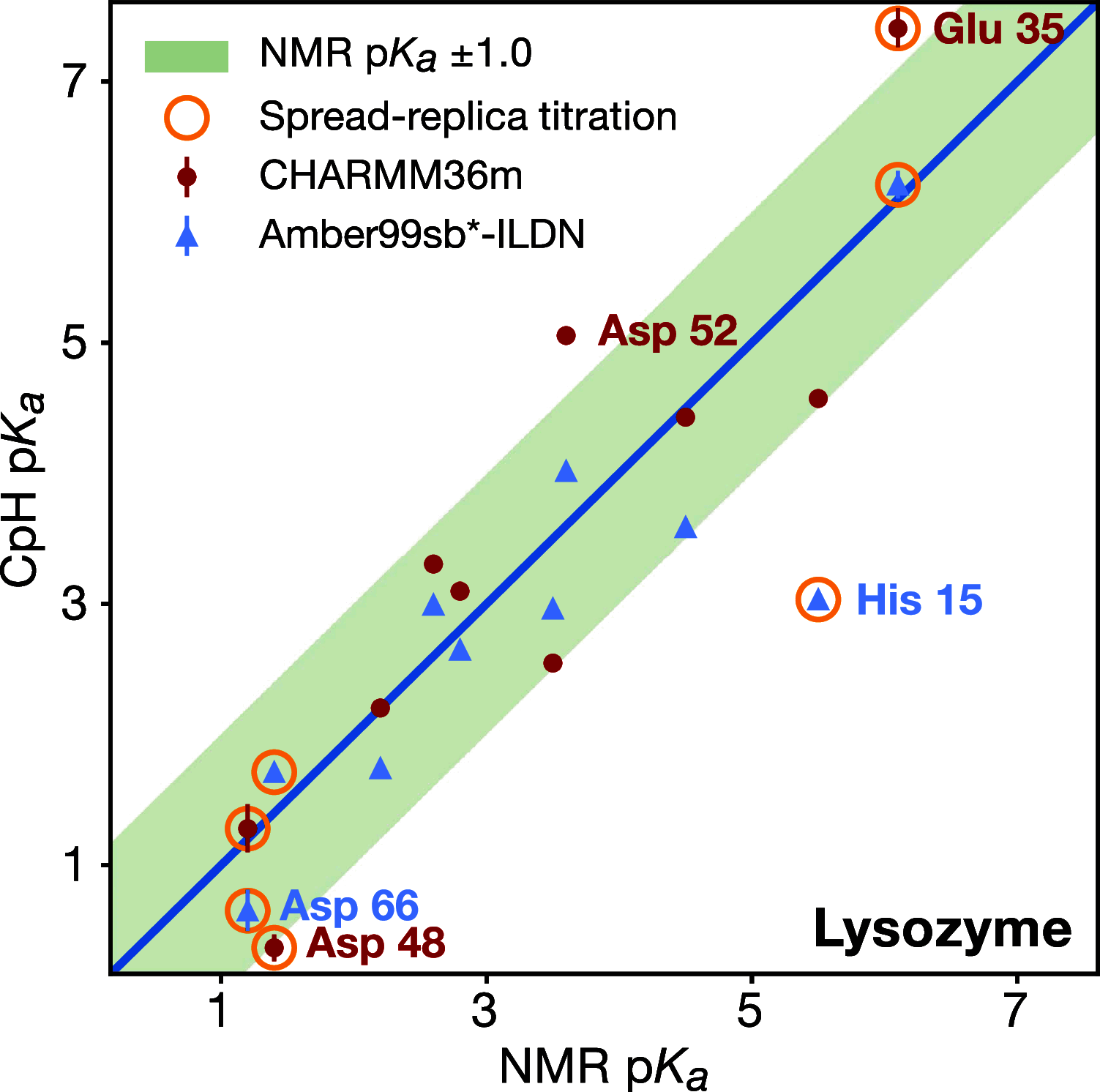
The structural dynamics of biological macromolecules, such as proteins, DNA/RNA, or complexes thereof, are strongly influenced by protonation changes of their typically many titratable groups, which explains their sensitivity to pH changes. Conversely, conformational and environmental changes of the biomolecule affect the protonation state of these groups. With few exceptions, conventional force field-based molecular dynamics (MD) simulations neither account for these effects nor do they allow for coupling to a pH buffer. Here, we present design decisions and applications of a rigorous Hamiltonian interpolation λ-dynamics constant pH method in GROMACS, which rests on GPU-accelerated Fast Multipole Method (FMM) electrostatics. Our implementation supports both CHARMM36m and Amber99sb*-ILDN force fields and is largely automated to enable seamless switching from regular MD to constant pH MD, involving minimal changes to the input files. Here, the first of two companion papers describes the underlying constant pH protocol and sample applications to several prototypical benchmark systems such as cardiotoxin V, lysozyme, and staphylococcal nuclease. Enhanced convergence is achieved through a new dynamic barrier height optimization method, and high pKa accuracy is demonstrated. We use Functional Mode Analysis (FMA) and Mutual Information (MI) to explore the complex intra- and intermolecular couplings between the protonation states of titratable groups as well as those between protonation states and conformational dynamics. We identify striking conformation-dependent pKa variations and unexpected inter-residue couplings. Conformation-protonation coupling is identified as a primary cause of the slow protonation convergence notorious to constant pH simulations involving multiple titratable groups, suggesting enhanced sampling methods to accelerate convergence.
Conflict of interest statement
The authors declare no competing financial interest.
Figures






Similar articles
-
Constant pH Simulation with FMM Electrostatics in GROMACS. (B) GPU Accelerated Hamiltonian Interpolation.J Chem Theory Comput. 2025 Feb 25;21(4):1787-1804. doi: 10.1021/acs.jctc.4c01319. Epub 2025 Feb 7. J Chem Theory Comput. 2025. PMID: 39919130 Free PMC article.
-
GPU-Accelerated All-Atom Particle-Mesh Ewald Continuous Constant pH Molecular Dynamics in Amber.J Chem Theory Comput. 2022 Dec 13;18(12):7510-7527. doi: 10.1021/acs.jctc.2c00586. Epub 2022 Nov 15. J Chem Theory Comput. 2022. PMID: 36377980 Free PMC article.
-
Constant-pH Molecular Dynamics Simulations for Large Biomolecular Systems.J Chem Theory Comput. 2017 Dec 12;13(12):5933-5944. doi: 10.1021/acs.jctc.7b00875. Epub 2017 Nov 22. J Chem Theory Comput. 2017. PMID: 29111720 Free PMC article.
-
Constant pH molecular dynamics simulations: Current status and recent applications.Curr Opin Struct Biol. 2022 Dec;77:102498. doi: 10.1016/j.sbi.2022.102498. Epub 2022 Nov 18. Curr Opin Struct Biol. 2022. PMID: 36410222 Free PMC article. Review.
-
Molecular dynamics simulation of proteins under high pressure: Structure, function and thermodynamics.Biochim Biophys Acta Gen Subj. 2020 Feb;1864(2):129395. doi: 10.1016/j.bbagen.2019.07.004. Epub 2019 Jul 11. Biochim Biophys Acta Gen Subj. 2020. PMID: 31302180 Review.
Cited by
-
KaMLs for Predicting Protein pKa Values and Ionization States: Are Trees All You Need?J Chem Theory Comput. 2025 Feb 11;21(3):1446-1458. doi: 10.1021/acs.jctc.4c01602. Epub 2025 Jan 30. J Chem Theory Comput. 2025. PMID: 39882632 Free PMC article.
-
KaMLs for Predicting Protein pK a Values and Ionization States: Are Trees All You Need?bioRxiv [Preprint]. 2025 Jan 30:2024.11.09.622800. doi: 10.1101/2024.11.09.622800. bioRxiv. 2025. Update in: J Chem Theory Comput. 2025 Feb 11;21(3):1446-1458. doi: 10.1021/acs.jctc.4c01602. PMID: 39605739 Free PMC article. Updated. Preprint.
-
Constant pH Simulation with FMM Electrostatics in GROMACS. (B) GPU Accelerated Hamiltonian Interpolation.J Chem Theory Comput. 2025 Feb 25;21(4):1787-1804. doi: 10.1021/acs.jctc.4c01319. Epub 2025 Feb 7. J Chem Theory Comput. 2025. PMID: 39919130 Free PMC article.
References
-
- Dlugosz M.; Antosiewicz J. M. Constant-pH molecular dynamics simulations: a test case of succinic acid. Chem. Phys. 2004, 302, 161–170. 10.1016/j.chemphys.2004.03.031. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
