MTA-Cooperative PRMT5 Inhibitors: Mechanism Switching Through Structure-Based Design
- PMID: 39919252
- PMCID: PMC11874000
- DOI: 10.1021/acs.jmedchem.4c01998
MTA-Cooperative PRMT5 Inhibitors: Mechanism Switching Through Structure-Based Design
Abstract
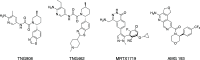
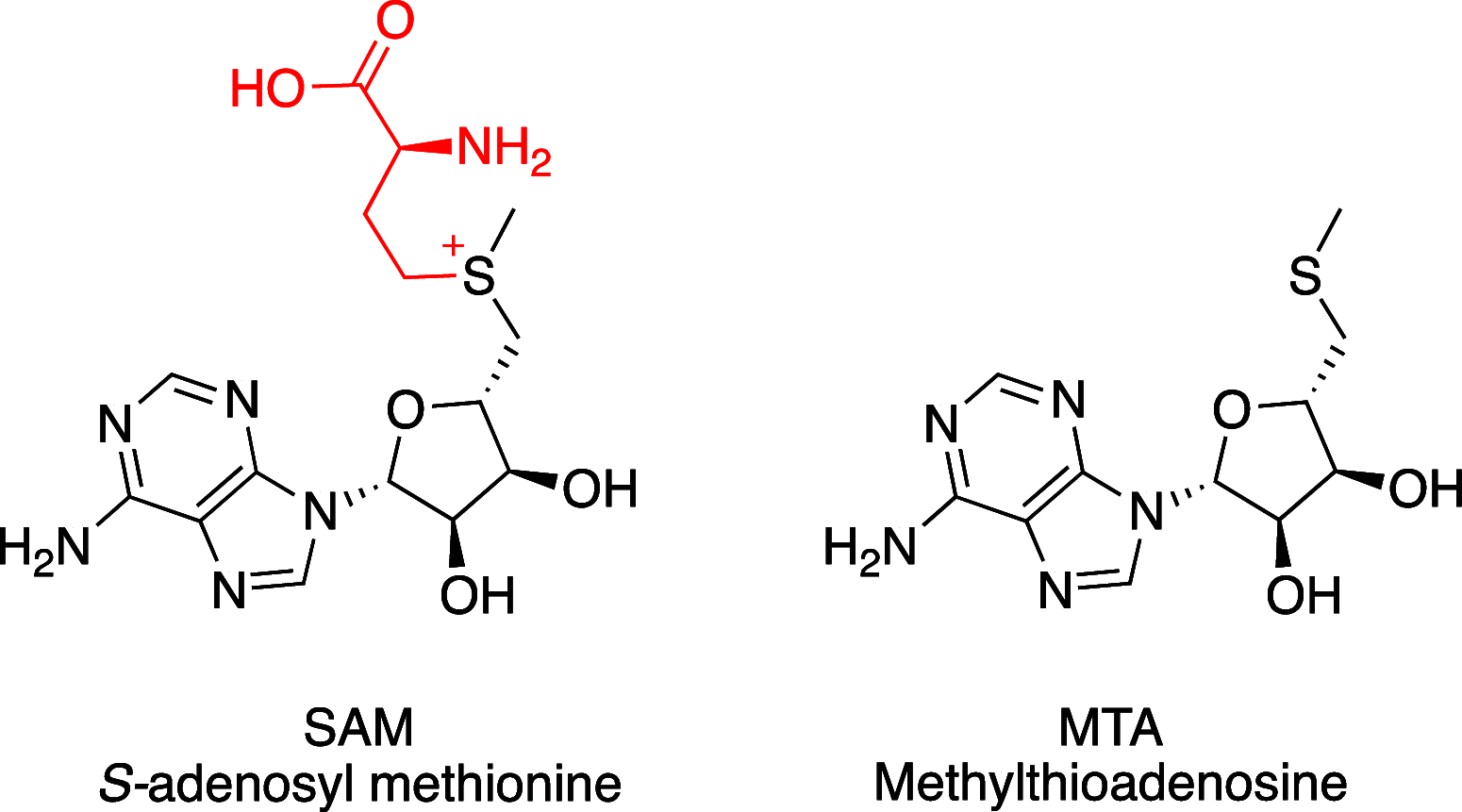
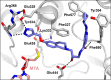
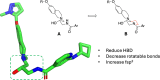
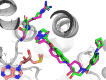
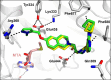
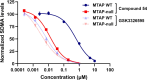
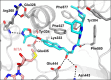
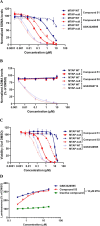
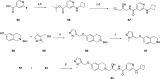
Deletion of the MTAP gene leads to accumulation of the substrate of the MTAP protein, methylthioadenosine (MTA). MTA binds PRMT5 competitively with S-adenosyl-l-methionine (SAM), and selective inhibition of the PRMT5•MTA complex relative to the PRMT5•SAM complex can lead to selective killing of cancer cells with MTAP deletion. Herein, we describe the discovery of novel compounds using structure-based drug design to switch the mechanism of binding of known, SAM-cooperative PRMT5 inhibitors to an MTA-cooperative binding mechanism by occupying the portion of the SAM binding pocket in PRMT5 that is unoccupied when MTA is bound and hydrogen bonding to Arg368, thereby allowing them to selectively target MTAP-deleted cancer cells.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Musiani D.; Bok J.; Massignani E.; Wu L.; Tabaglio T.; Ippolito M. R.; Cuomo A.; Ozbek U.; Zorgati H.; Ghoshdastider U.; Robinson R. C.; Guccione E.; Bonaldi T. Proteomics Profiling of Arginine Methylation Defines PRMT5 Substrate Specificity. Sci. Signaling 2019, 12 (575), eaat838810.1126/scisignal.aat8388. - DOI - PubMed
-
- Chan-Penebre E.; Kuplast K. G.; Majer C. R.; Boriack-Sjodin P. A.; Wigle T. J.; Johnston L. D.; Rioux N.; Munchhof M. J.; Jin L.; Jacques S. L.; West K. A.; Lingaraj T.; Stickland K.; Ribich S. A.; Raimondi A.; Scott M. P.; Waters N. J.; Pollock R. M.; Smith J. J.; Barbash O.; Pappalardi M.; Ho T. F.; Nurse K.; Oza K. P.; Gallagher K. T.; Kruger R.; Moyer M. P.; Copeland R. A.; Chesworth R.; Duncan K. W. A Selective Inhibitor of PRMT5 with in Vivo and in Vitro Potency in MCL Models. Nat. Chem. Biol. 2015, 11 (6), 432–437. 10.1038/nchembio.1810. - DOI - PubMed
-
- Brehmer D.; Beke L.; Wu T.; Millar H. J.; Moy C.; Sun W.; Mannens G.; Pande V.; Boeckx A.; van Heerde E.; et al. Discovery and Pharmacological Characterization of JNJ-64619178, a Novel Small Molecule Inhibitor of PRMT5 with Potent Anti-Tumor Activity. Mol. Cancer Ther. 2021, 20 (12), 2317–2328. 10.1158/1535-7163.mct-21-0367. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

