WNT11 Promotes immune evasion and resistance to Anti-PD-1 therapy in liver metastasis
- PMID: 39920102
- PMCID: PMC11806061
- DOI: 10.1038/s41467-025-56714-z
WNT11 Promotes immune evasion and resistance to Anti-PD-1 therapy in liver metastasis
Abstract
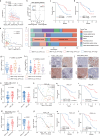
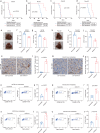
Liver metastasis (LM) poses a significant challenge in cancer treatment, with limited available therapeutic options and poor prognosis. Understanding the dynamics of tumor microenvironment (TME) and immune interactions is crucial for developing effective treatments. We find that WNT11 promoted CD8+ T-cell exclusion and suppression, which was correlated with poor prognosis in LM. Mechanistically, WNT11-overexpressing tumor cells directly reduce CD8+ T-cell recruitment and activity by decreasing CXCL10 and CCL4 expression through CAMKII-mediated β-catenin/AFF3 downregulation. WNT11-overexpressing tumor cells promote immunosuppressive macrophage polarization by inducing IL17D expression via the CAMKII/NF-κB pathway, which result in CD8+ T-cell suppression. Moreover, CAMKII inhibition increases the efficacy of anti-PD-1 therapy in mouse model of LM. Serum expression of WNT11 is identified as a potential minimally invasive biomarker in the management of colorectal cancer-LM with immunotherapy. Our findings highlight WNT11/CAMKII axis as a critical regulator of the TME and a promising target for immunotherapy in patients with LM.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Tsilimigras, D. I. et al. Liver metastases. Nat. Rev. Dis. Prim.7, 1–23 (2021). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

