Orphan G protein-coupled receptor GPRC5B controls macrophage function by facilitating prostaglandin E receptor 2 signaling
- PMID: 39920161
- PMCID: PMC11805951
- DOI: 10.1038/s41467-025-56713-0
Orphan G protein-coupled receptor GPRC5B controls macrophage function by facilitating prostaglandin E receptor 2 signaling
Abstract
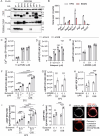
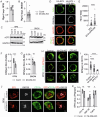
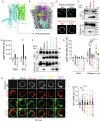
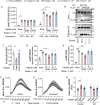
Macrophages express numerous G protein-coupled receptors (GPCRs) that regulate adhesion, migration, and activation, but the function of orphan receptor GPRC5B in macrophages is unknown. Both resident peritoneal and bone marrow-derived macrophages from myeloid-specific GPRC5B-deficient mice show increased migration and phagocytosis, resulting in improved bacterial clearance in a peritonitis model. In other models such as myocardial infarction, increased myeloid cell recruitment has adverse effects. Mechanistically, we found that GPRC5B physically interacts with GPCRs of the prostanoid receptor family, resulting in enhanced signaling through the prostaglandin E receptor 2 (EP2). In GPRC5B-deficient macrophages, EP2-mediated anti-inflammatory effects are diminished, resulting in hyperactivity. Using in silico modelling and docking, we identify residues potentially mediating GPRC5B/EP2 dimerization and show that their mutation results in loss of GPRC5B-mediated facilitation of EP2 signaling. Finally, we demonstrate that decoy peptides mimicking the interacting sequence are able to reduce GPRC5B-mediated facilitation of EP2-induced cAMP signaling in macrophages.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Varol, C., Mildner, A. & Jung, S. Macrophages: development and tissue specialization. Annu Rev. Immunol.33, 643–675 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

