An Integrated Database for Exploring Alternative Promoters in Animals
- PMID: 39920194
- PMCID: PMC11805906
- DOI: 10.1038/s41597-025-04548-1
An Integrated Database for Exploring Alternative Promoters in Animals
Abstract
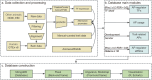
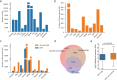
Alternative promoter (AP) events, as a major pre-transcriptional mechanism, can initiate different transcription start sites to generate distinct mRNA isoforms and regulate their expression. At present, hundreds of thousands of APs have been identified across human tissues, and a considerable number of APs have been demonstrated to be associated with complex traits and diseases. Recent researches have also proven important effects of APs on animals. However, the landscape of APs in animals has not been fully recognized. In this study, 102,349 AP profiles from 23,077 samples across 12 species were systematically characterized. We further identified tissue-specific APs and investigated trait-related promoters among various species. In addition, we analyzed the associations between APs and enhancer RNAs (eRNA)/transcription factors (TF) as a means of identifying potential regulatory factors. Integrating these findings, we finally developed Animal-APdb, a database for the searching, browsing, and downloading of information related to Animal APs. Animal-APdb is expected to serve as a valuable resource for exploring the functions and mechanisms of APs in animals.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
Animal-eRNAdb: a comprehensive animal enhancer RNA database.Nucleic Acids Res. 2022 Jan 7;50(D1):D46-D53. doi: 10.1093/nar/gkab832. Nucleic Acids Res. 2022. PMID: 34551433 Free PMC article.
-
Animal-APAdb: a comprehensive animal alternative polyadenylation database.Nucleic Acids Res. 2021 Jan 8;49(D1):D47-D54. doi: 10.1093/nar/gkaa778. Nucleic Acids Res. 2021. PMID: 32986825 Free PMC article.
-
CrusTF: a comprehensive resource of transcriptomes for evolutionary and functional studies of crustacean transcription factors.BMC Genomics. 2017 Nov 25;18(1):908. doi: 10.1186/s12864-017-4305-2. BMC Genomics. 2017. PMID: 29178828 Free PMC article.
-
Promoter or enhancer, what's the difference? Deconstruction of established distinctions and presentation of a unifying model.Bioessays. 2015 Mar;37(3):314-23. doi: 10.1002/bies.201400162. Epub 2014 Dec 2. Bioessays. 2015. PMID: 25450156 Review.
-
Regulation of gene expression by alternative promoters.FASEB J. 1996 Mar;10(4):453-60. FASEB J. 1996. PMID: 8647344 Review.
References
-
- Ayoubi, T. A. & Van De Ven, W. J. Regulation of gene expression by alternative promoters. FASEB J10, 453–460 (1996). - PubMed
-
- Davuluri, R. V., Suzuki, Y., Sugano, S., Plass, C. & Huang, T. H. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet24, 167–177, 10.1016/j.tig.2008.01.008 (2008). - PubMed
-
- Bieberstein, N. I., Carrillo Oesterreich, F., Straube, K. & Neugebauer, K. M. First exon length controls active chromatin signatures and transcription. Cell Rep2, 62–68, 10.1016/j.celrep.2012.05.019 (2012). - PubMed
-
- Schibler, U. & Sierra, F. Alternative promoters in developmental gene expression. Annu Rev Genet21, 237–257, 10.1146/annurev.ge.21.120187.001321 (1987). - PubMed
-
- Maqbool, M. A. et al. Alternative Enhancer Usage and Targeted Polycomb Marking Hallmark Promoter Choice during T Cell Differentiation. Cell Rep32, 108048, 10.1016/j.celrep.2020.108048 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2021CFB404/Natural Science Foundation of Hubei Province (Hubei Provincial Natural Science Foundation)
- 2662022XXYJ008/Huazhong Agricultural University (HZAU)
- 11041810351/Huazhong Agricultural University (HZAU)
- 31970644/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Miscellaneous

