Genome-wide association studies dissect the genetic architecture of seed and yield component traits in cowpea (Vigna unguiculata L. Walp)
- PMID: 39920462
- PMCID: PMC12005157
- DOI: 10.1093/g3journal/jkaf024
Genome-wide association studies dissect the genetic architecture of seed and yield component traits in cowpea (Vigna unguiculata L. Walp)
Abstract
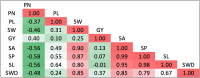
The identification of loci related to seed and yield component traits in cowpea constitutes a key step for improvement through marker-assisted selection (MAS). Furthermore, seed morphology has an impact on industrial processing and influences consumer and farmer preferences. In this study, we performed genome-wide association studies (GWAS) on a mini-core collection of cowpea to dissect the genetic architecture and detect genomic regions associated with seed morphological traits and yield components. Phenotypic data were measured both manually and by high-throughput image-based approaches to test associations with 41,533 single nucleotide polymorphism markers using the FarmCPU model. From genome-associated regions, we also investigated putative candidate genes involved in the variation of the phenotypic traits. We detected 42 marker-trait associations for pod length and 100-seed weight, length, width, perimeter, and area of the seed. Candidate genes encoding leucine-rich repeat-containing (LRR) and F-box proteins, known to be associated with seed size, were identified; in addition, we identified candidate genes encoding PPR (pentatricopeptide repeat) proteins, recognized to have an important role in seed development in several crops. Our findings provide insights into natural variation in cowpea for yield-related traits and valuable information for MAS breeding strategies in this and other closely related crops.
Keywords: Vigna unguiculata; GWAS; grain yield; image-based phenotyping; seed morphology.
© The Author(s) 2025. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The author(s) declare no conflict of interest.
Figures

Similar articles
-
A genome-wide association and meta-analysis reveal regions associated with seed size in cowpea [Vigna unguiculata (L.) Walp].Theor Appl Genet. 2019 Nov;132(11):3079-3087. doi: 10.1007/s00122-019-03407-z. Epub 2019 Jul 31. Theor Appl Genet. 2019. PMID: 31367839 Free PMC article.
-
Identification of Candidate Genes Controlling Black Seed Coat and Pod Tip Color in Cowpea (Vigna unguiculata [L.] Walp).G3 (Bethesda). 2018 Oct 3;8(10):3347-3355. doi: 10.1534/g3.118.200521. G3 (Bethesda). 2018. PMID: 30143525 Free PMC article.
-
Genome-wide association studies dissect the genetic architecture of seed shape and size in common bean.G3 (Bethesda). 2022 Apr 4;12(4):jkac048. doi: 10.1093/g3journal/jkac048. G3 (Bethesda). 2022. PMID: 35218340 Free PMC article.
-
Assessing the genetic diversity of cowpea [Vigna unguiculata (L.) Walp.] germplasm collections using phenotypic traits and SNP markers.BMC Genet. 2020 Sep 18;21(1):110. doi: 10.1186/s12863-020-00914-7. BMC Genet. 2020. PMID: 32948123 Free PMC article.
-
Introgression Breeding in Cowpea [Vigna unguiculata (L.) Walp.].Front Plant Sci. 2020 Sep 16;11:567425. doi: 10.3389/fpls.2020.567425. eCollection 2020. Front Plant Sci. 2020. PMID: 33072144 Free PMC article. Review.
References
-
- Abebe BK, Alemayehu MT. 2022. A review of the nutritional use of cowpea (Vigna unguiculata L. Walp) for human and animal diets. J Agric Food Res. 10:100383. doi:10.1016/j.jafr.2022.100383. - DOI
-
- Agbicodo EM, Fatokun CA, Muranaka S, Visser RG, Linden Van Der CG. 2009. Breeding drought tolerant cowpea: constraints, accomplishments, and future prospects. Euphytica. 167:353–370. doi:10.1007/s10681-009-9893-8 - DOI
-
- Aliyu OM, Makinde BO. 2016. Phenotypic analysis of seed yield and yield components in cowpea (Vigna unguiculata L., Walp). Plant Breed Biotechnol. 4(2):252–261. doi:10.9787/PBB.2016.4.2.252. - DOI
-
- Andrade MHML, Ferreira RCU, Fernandes CCF, Sipowicz P, Rios EF. 2023. Single and multi-trait genome-wide association studies identify genomic regions associated with phenological traits in cowpea. Crop Sci. 63:3443–3456. doi:10.1002/csc2.21079. - DOI
-
- Araújo JPP. 1988. Melhoramento do caupi no Brasil. In: de Araújo JPP, Watt EE, editors. O caupi no Brasil. Brasília (DF): IITA; EMBRAPA. p. 249–283.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
