Loss of MEF2C function by enhancer mutation leads to neuronal mitochondria dysfunction and motor deficits in mice
- PMID: 39920775
- PMCID: PMC11806887
- DOI: 10.1186/s13024-024-00792-y
Loss of MEF2C function by enhancer mutation leads to neuronal mitochondria dysfunction and motor deficits in mice
Abstract
Background: Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disorder characterized by the loss of both upper and lower motor neurons, leading to progressive paralysis. Both genetic alterations and epigenetic modifications contribute to neuronal dysfunction in the pathogenesis of ALS. However, the mechanism behind genetic mutations in the non-coding region of genes that affect epigenetic modifications remains unclear.
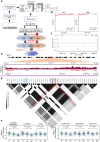
Methods: Convolutional neural network was used to identify an ALS-associated SNP located in the intronic region of MEF2C (rs304152), residing in a putative enhancer element. To examine the alteration of MEF2C transcription by the SNP, we generated HEK293T cells carrying the major or minor allele by CRISPR-Cas9. To verify the role of MEF2C-knockdown (MEF2C-KD) in mice, we developed AAV expressing shRNA for MEF2C based on AAV-U6 promoter vector. Neuropathological alterations of MEF2C-KD mice with mitochondrial dysfunction and motor neuronal damage were observed by confocal microscopy and transmission electron microscope (TEM). Behavioral changes of mice were examined through longitudinal study by tail suspension, inverted grid test and automated gait analysis.
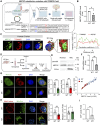
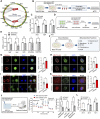
Results: Here, we show that enhancer mutation of MEF2C reduces own gene expression and consequently impairs mitochondrial function in motor neurons. MEF2C localizes and binds to the mitochondria DNA, and directly modulates mitochondria-encoded gene expression. CRISPR/Cas-9-induced mutation of the MEF2C enhancer decreases expression of mitochondria-encoded genes. Moreover, MEF2C mutant cells show reduction of mitochondrial membrane potential, ATP level but elevation of oxidative stress. MEF2C deficiency in the upper and lower motor neurons of mice impairs mitochondria-encoded genes, and leads to mitochondrial metabolic disruption and progressive motor behavioral deficits.
Conclusions: Together, MEF2C dysregulation by the enhancer mutation leads to mitochondrial dysfunction and oxidative stress, which are prevalent features in motor neuronal damage and ALS pathogenesis. This genetic and epigenetic crosstalk mechanism provides insights for advancing our understanding of motor neuron disease and developing effective treatments.
Keywords: Epigenetics; MEF2C; Mitochondria; Motor neuron; Single nucleotide polymorphism (SNP).
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent participate: All animal (mouse) work and experiments in this study were approved by KIST Institutional Animal Care and Use Committee (IACUC). Consent for publication: All authors have consented for the publication of manuscript. Competing interests: The authors report no competing interests.
Figures



Update of
-
Loss of MEF2C function by enhancer mutation leads to neuronal mitochondria dysfunction and motor deficits in mice.bioRxiv [Preprint]. 2024 Jul 17:2024.07.15.603186. doi: 10.1101/2024.07.15.603186. bioRxiv. 2024. Update in: Mol Neurodegener. 2025 Feb 07;20(1):16. doi: 10.1186/s13024-024-00792-y. PMID: 39071309 Free PMC article. Updated. Preprint.
References
-
- Haukedal H, Freude K. Implications of Microglia in Amyotrophic lateral sclerosis and Frontotemporal Dementia. J Mol Biol. 2019;431(9):1818–29. - PubMed
-
- Xi Z, Rainero I, Rubino E, Pinessi L, Bruni AC, Maletta RG, Nacmias B, Sorbi S, Galimberti D, Surace EI, et al. Hypermethylation of the CpG-island near the C9orf72 G₄C₂-repeat expansion in FTLD patients. Hum Mol Genet. 2014;23(21):5630–7. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

