Diamonds in the rif: Alignment-free comparative genomics analysis reveals strain-transcendent Plasmodium falciparum antigens amidst extensive genetic diversity
- PMID: 39920908
- PMCID: PMC12032969
- DOI: 10.1016/j.meegid.2025.105725
Diamonds in the rif: Alignment-free comparative genomics analysis reveals strain-transcendent Plasmodium falciparum antigens amidst extensive genetic diversity
Abstract
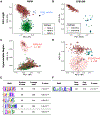
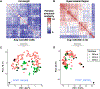
The repetitive interspersed family (rif) and subtelomeric variable open reading frames (stevor) are highly diverse multi-gene families in the malaria parasite Plasmodium falciparum. Embedded on the surface of infected erythrocytes, RIFIN and STEVOR proteins are involved in cytoadherence and immune evasion, but the extent of family-wide sequence diversity across strains has yet to be comprehensively investigated in light of improved resolution of the subtelomeric genome sequences. Using a k-mer frequency approach, we analyzed long-read genomic sequence data from 18 geographically diverse P. falciparum genome assemblies, including lab strains and clinical isolates. We hypothesized that k-mer sequence comparison can identify existing RIFIN and STEVOR subgroups, identify novel subgroups, and generate more robust and reliable estimates of family-wide sequence diversity. Full-length RIFIN and STEVOR proteins shared on average 49.5% and 61.1% amino acid k-mer similarity, respectively, which fell to 25.1% and 20% in the hypervariable regions alone. Despite this diversity, we identified 11 RIFINs and five STEVORs that were conserved across strains above expected thresholds. A subset of these strain-transcendent genes was similar and syntenic to genes in related Plasmodium species, suggesting an ancient origin. Additionally, in silico structural predictions from AlphaFold showed that three-dimensional structures of RIFIN receptor-binding regions were more conserved than their sequences suggested. Evolutionarily constrained RIFINs and STEVORs may have critical functions in parasite survival or pathogenesis. This study provides a framework for investigating diversity in highly variable multi-gene families and highlights the potential of strain-transcendent RIFIN and STEVOR proteins as vaccine candidates.
Keywords: Comparative genomics; Malaria; Plasmodium falciparum; RIFIN; STEVOR; Variant surface antigens.
Copyright © 2025 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest Research Support This research received no external financial or non-financial support. Relationships There are no additional relationships to disclose. Patents and Intellectual Property There are no patents to disclose. Other Activities There are no additional activities to disclose.
Figures



Similar articles
-
RSpred, a set of Hidden Markov Models to detect and classify the RIFIN and STEVOR proteins of Plasmodium falciparum.BMC Genomics. 2011 Feb 18;12:119. doi: 10.1186/1471-2164-12-119. BMC Genomics. 2011. PMID: 21332983 Free PMC article.
-
Whole genome sequencing and microsatellite analysis of the Plasmodium falciparum E5 NF54 strain show that the var, rifin and stevor gene families follow Mendelian inheritance.Malar J. 2018 Oct 22;17(1):376. doi: 10.1186/s12936-018-2503-2. Malar J. 2018. PMID: 30348135 Free PMC article.
-
Method for deletion of variant surface antigen genes at subtelomeric region of Plasmodium falciparum.Parasitol Int. 2025 Dec;109:103112. doi: 10.1016/j.parint.2025.103112. Epub 2025 Jun 23. Parasitol Int. 2025. PMID: 40562206
-
Plasmodium falciparum genetic diversity and multiplicity of infection based on msp-1, msp-2, glurp and microsatellite genetic markers in sub-Saharan Africa: a systematic review and meta-analysis.Malar J. 2024 Apr 8;23(1):97. doi: 10.1186/s12936-024-04925-y. Malar J. 2024. PMID: 38589874 Free PMC article.
-
Primaquine or other 8-aminoquinoline for reducing P. falciparum transmission.Cochrane Database Syst Rev. 2014 Jun 30;(6):CD008152. doi: 10.1002/14651858.CD008152.pub3. Cochrane Database Syst Rev. 2014. Update in: Cochrane Database Syst Rev. 2015 Feb 19;(2):CD008152. doi: 10.1002/14651858.CD008152.pub4. PMID: 24979199 Free PMC article. Updated.
References
-
- Albrecht L, Merino EF, Hoffmann EHE, Ferreira MU, de Mattos Ferreira RG, Osakabe AL, Dalla Martha RC, Ramharter M, Durham AM, Ferreira JE, del Portillo HA, Wunderlich G, 2006. Extense variant gene family repertoire overlap in Western Amazon plasmodium falciparum isolates. Mol. Biochem. Parasitol. 150 (2), 157–165. 10.1016/j.molbiopara.2006.07.007. - DOI - PubMed
-
- Amos B, Aurrecoechea C, Barba M, Barreto A, Basenko EY, Bażant W, Belnap R, Blevins AS, Böhme U, Brestelli J, Brunk BP, Caddick M, Callan D, Campbell L, Christensen MB, Christophides GK, Crouch K, Davis K, DeBarry J, Zheng J, 2022. VEuPathDB: the eukaryotic pathogen, vector and host bioinformatics resource center. Nucleic Acids Res. 50 (D1), D898–D911. 10.1093/nar/gkab929. - DOI - PMC - PubMed
-
- Benavente ED, Oresegun DR, de Sessions PF, Walker EM, Roper C, Dombrowski JG, de Souza RM, Marinho CRF, Sutherland CJ, Hibberd ML, Mohareb F, Baker DA, Clark TG, Campino S, 2018. Global genetic diversity of var2csa in plasmodium falciparum with implications for malaria in pregnancy and vaccine development. Sci. Rep. 8 (1). 10.1038/s41598-018-33767-3. Article 1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

