Longitudinal MRI-Driven Multi-Modality Approach for Predicting Pathological Complete Response and B Cell Infiltration in Breast Cancer
- PMID: 39921294
- PMCID: PMC11948082
- DOI: 10.1002/advs.202413702
Longitudinal MRI-Driven Multi-Modality Approach for Predicting Pathological Complete Response and B Cell Infiltration in Breast Cancer
Abstract
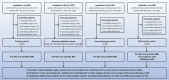
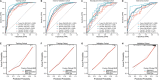
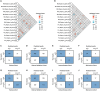
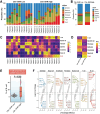
Accurately predicting pathological complete response (pCR) to neoadjuvant treatment (NAT) in breast cancer remains challenging due to tumor heterogeneity. This study enrolled 2279 patients across 12 centers and develops a novel multi-modality model integrating longitudinal magnetic resonance imaging (MRI) spatial habitat radiomics, transcriptomics, and single-cell RNA sequencing for predicting pCR. By analyzing tumor subregions on multi-timepoint MRI, the model captures dynamic intra-tumoral heterogeneity during NAT. It shows superior performance over traditional radiomics, with areas under the curve of 0.863, 0.813, and 0.888 in the external validation, immunotherapy, and multi-omics cohorts, respectively. Subgroup analysis shows its robustness across varying molecular subtypes and clinical stages. Transcriptomic and single-cell RNA sequencing analysis reveals that high model scores correlate with increased immune activity, notably elevated B cell infiltration, indicating the biological basis of the imaging model. The integration of imaging and molecular data demonstrates promise in spatial habitat radiomics to monitor dynamic changes in tumor heterogeneity during NAT. In clinical practice, this study provides a noninvasive tool to accurately predict pCR, with the potential to guide treatment planning and improve breast-conserving surgery rates. Despite promising results, the model requires prospective validation to confirm its utility across diverse patient populations and clinical settings.
Keywords: artificial intelligence; breast cancer; medical imaging; multi‐omics analysis; neoadjuvant treatment.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Byrd D. R., Brierley J. D., Baker T. P., Sullivan D. C., Gress D. M., Ca‐Cancer J. Clin. 2021, 71, 140. - PubMed
-
- Cortazar P., Zhang L., Untch M., Mehta K., Costantino J. P., Wolmark N., Bonnefoi H., Cameron D., Gianni L., Valagussa P., Swain S. M., Prowell T., Loibl S., Wickerham D. L., Bogaerts J., Baselga J., Perou C., Blumenthal G., Blohmer J., Mamounas E. P., Bergh J., Semiglazov V., Justice R., Eidtmann H., Paik S., Piccart M., Sridhara R., Fasching P. A., Slaets L., Tang S., et al., Lancet 2014, 384, 164. - PubMed
-
- Parker J. S., Mullins M., Cheang M. C. U., Leung S., Voduc D., Vickery T., Davies S., Fauron C., He X., Hu Z., Quackenbush J. F., Stijleman I. J., Palazzo J., Marron J. S., Nobel A. B., Mardis E., Nielsen T. O., Ellis M. J., Perou C. M., Bernard P. S., J. Clin. Oncol. 2023, 41, 4192. - PubMed
MeSH terms
Grants and funding
- 82171898/National Natural Science Foundation of China
- DFJHBF202109/High-level Hospital Construction Project of Guangdong Provincial People's Hospital
- 2021A1515010737/Basic and Applied Basic Research Foundation of Guangdong Province
- 2023A1515010222/Basic and Applied Basic Research Foundation of Guangdong Province
- 202002030236/Guangzhou Science and Technology Project
- 32070676/Natural Science Foundation Council of China
- 20210701181316106/AKP/Macao Science and Technology Development Fund
- YXJL-2020-0941-0758/Beijing Medical Award Foundation
- KC2022-ZZ-0091-5/Beijing Science and Technology Innovation Medical Development Foundation
- WKZX2023CX110002/Development Cancer for Medical Science and Technology National Health Commission of the People's Republic of China
- cphcf-2022-058/Beijing Life Oasis Public Service Center
- JW2023001/Changzhou Medical Talents Project for Domestic and Foreign Training
LinkOut - more resources
Full Text Sources
Medical
