Transcriptomic Profiling Unveils EDN3+ Meningeal Fibroblasts as Key Players in Sturge-Weber Syndrome Pathogenesis
- PMID: 39921427
- PMCID: PMC12061316
- DOI: 10.1002/advs.202408888
Transcriptomic Profiling Unveils EDN3+ Meningeal Fibroblasts as Key Players in Sturge-Weber Syndrome Pathogenesis
Abstract
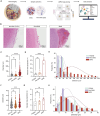
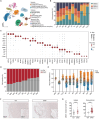
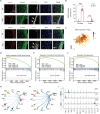
Sturge-Weber syndrome (SWS) is characterized by leptomeningeal vascular malformation, resulting in significant risks of life-threatening seizures and strokes. The current absence of specific treatments underscores the need to define the molecular and cellular mechanisms that drive the progression of SWS. Here, the transcriptome of 119 446 cells isolated from both malformed tissues and peri-lesion tissues from the brains of patients with SWS is examined. This comprehensive analysis finds a complex landscape of cell heterogeneity and distinct cell substate associated with the evolution of this disease are revealed. Notably, a unique fibroblast cluster and molecular mechanism are identified that contribute to the development of SWS. These findings not only expand the understanding of SWS but also open up promising avenues for therapeutic interventions.
Keywords: Sturge‐Weber syndrome; cerebrovasculature; meningeal fibroblasts; single‐cell RNA sequencing.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
