Selection Increases Mitonuclear DNA Discordance but Reconciles Incompatibility in African Cattle
- PMID: 39921600
- PMCID: PMC11879056
- DOI: 10.1093/molbev/msaf039
Selection Increases Mitonuclear DNA Discordance but Reconciles Incompatibility in African Cattle
Abstract
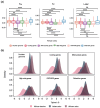
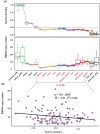
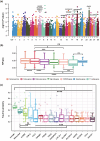
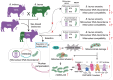
Mitochondrial function relies on the coordinated interactions between genes in the mitochondrial DNA and nuclear genomes. Imperfect interactions following mitonuclear incompatibility may lead to reduced fitness. Mitochondrial DNA introgressions across species and populations are common and well documented. Various strategies may be expected to reconcile mitonuclear incompatibility in hybrids or admixed individuals. African admixed cattle (Bos taurus × B. indicus) show sex-biased admixture, with taurine (B. taurus) mitochondrial DNA and a nuclear genome predominantly of humped zebu (B. indicus). Here, we leveraged local ancestry inference approaches to identify the ancestry and distribution patterns of nuclear functional genes associated with the mitochondrial oxidative phosphorylation process in the genomes of African admixed cattle. We show that most of the nuclear genes involved in mitonuclear interactions are under selection and of humped zebu ancestry. Variations in mitochondrial DNA copy number may have contributed to the recovery of optimal mitochondrial function following admixture with the regulation of gene expression, alleviating or nullifying mitochondrial dysfunction. Interestingly, some nuclear mitochondrial genes with enrichment in taurine ancestry may have originated from ancient African aurochs (B. primigenius africanus) introgression. They may have contributed to the local adaptation of African cattle to pathogen burdens. Our study provides further support and new evidence showing that the successful settlement of cattle across the continent was a complex mechanism involving adaptive introgression, mitochondrial DNA copy number variation, regulation of gene expression, and selection of ancestral mitochondria-related genes.
Keywords: African cattle; admixture; mitonuclear DNA discordance; mitonuclear interactions; mtDNA copy numbers; purifying selection.
© The Author(s) 2025. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of Interest: The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

