Molecular characterization of pregnancy-associated breast cancer and insights on timing from GEICAM-EMBARCAM study
- PMID: 39922815
- PMCID: PMC11807221
- DOI: 10.1038/s41523-025-00718-x
Molecular characterization of pregnancy-associated breast cancer and insights on timing from GEICAM-EMBARCAM study
Abstract
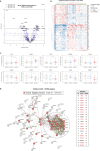
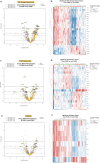
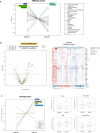
Pregnancy-associated breast cancer (PABC), diagnosed during or shortly after pregnancy, is a challenging entity with an aggressive biology and poor prognosis. This study analyzed the clinicopathological characteristics and gene expression profile of 33 PABC and 26 non-PABC patients using the nCounter BC360 Panel (NanoString). Notably, PABC showed a higher prevalence of basal-like tumors than non-PABC (48.48% vs 15.38%, p = 0.012) and displayed 73 differentially expressed genes (e.g., DEPDC1, CCNA2, PSAT1, CDKN3, and FAM83D), enriched in DNA repair and cell proliferation pathways. Through the PPI network, we also identified a cluster of cell-cycle regulation genes like MYC, FOXM1, or PTEN. Interestingly, differences emerged when comparing patients diagnosed during gestation (PABC-GS) and the postpartum period (PABC-PP), with PABC-PP showing increased expression of immune-related genes, including PD-1, and greater immune cell infiltration (Tregs, macrophages, neutrophils, B-cells). These findings suggest an enhanced proliferative capacity and impaired DNA repair in PABC, and underscore the role of immune infiltration in postpartum cases; providing insights into its aggressive nature and potential targets.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: B.B. declares having received fees for medical education in a consulting or advisory role with Lilly, Pfizer, MSD, Pierr Fabre, Astra Zeneca, and Gilead; participated in a speakers’ bureau with Roche, MSD, Daichii, Sankio Astra Zeneca, Novartis, Lilly, Gilead, Seagen; travel accommodation by Pfizer and Gilead. Y.J.G. declares having received speakers´ honoraria from Roche, Novartis, Lilly, Daichii, and AstraZeneca; travel and training grants from Roche, Novartis, Pfizer, Daichii, Gilead, and Lilly. A.F.A. declares Honoraria or consultation fees from GSK, MSD, AstraZeneca, Pharma & Go, EISAI, Lilly, Pfizer, Novartis, and Pierre Fabre; travel/accommodation expenses from GSK, MSD, AstraZeneca, and Pfizer; speakers bureau participation with GSK, MSD, AstraZeneca, EISAI, Novartis and Pierre Fabre. F.M. declares consulting/advisory role with Novartis, Pfizer, AstraZeneca, MSD, Daiichi Sankyo, and Seagen; speakers’ bureau from Pfizer and Novartis; research funding from Pfizer; travel/accommodations expenses by Pfizer, Novartis and Gilead. M.H.L.-C. declares having received speaker honoraria from Daichii, Novartis & Pierre Fabre; and travel and training grants from Roche & Novartis. I.B. declares having research funding from Agendia, AstraZeneca, Lilly, Pfizer, and Roche; honoraria as a medical monitor from Medical Scientia Innovation Research (MEDSIR); honoraria and advisor collaboration from AstraZeneca, Bristol-Myers Squibb, Celgene, Daiichi-Sankyo, Eisai, Gilead, Grünenthal, GSK, Jazz Pharmaceutical, Lilly, MSD, Novartis, Pfizer, Pierre-Fabre, Roche, Seagen and Veracyte; travel and meeting attendance grants from AstraZeneca, Bristol-Myers Squibb, Daiichi-Sankyo, Gilead, Lilly, Novartis, Pfizer, Pierre-Fabre and Roche. A.G.-Z. declares institutional grant from Pfizer; advisory role honoraria from AstraZeneca, Novartis, MSD, Pierre-Fabre, Exact Science, Menarini-Stemline and Daichy Sankyo; travel grants from Roche, Novartis, Pfizer, Gilead and Pierre Fabre; speaker Bureau/Expert testimony with Roche, AstraZeneca, Daichi-Sankyo, Novartis, MSD, Pfizer and Menarini-Stemline. J.H.-R. declares having received research grants from Roche and Pfizer; consulting/advisory fees from AstraZeneca, Amgen, Roche/Genentech, Novartis, Eli Lilly, and Pfizer; speakers’ honoraria from AstraZeneca, Lilly, Amgen, Roche/Genentech, Novartis, and Pfizer. All remaining authors have declared no conflicts of interest.
Figures




References
-
- Peccatori, F. A. et al. Cancer, pregnancy and fertility: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann.Oncol. 10.1093/ANNONC/MDT199 (2013). - PubMed
-
- Vinatier, E., Merlot, B., Poncelet, E., Collinet, P. & Vinatier, D. Breast cancer during pregnancy. Eur. J. Obstet. Gynecol. Reprod. Biol.147, 9–14 (2009). - PubMed
-
- Johansson, A. L. V. & Stensheim, H. Epidemiology of pregnancy-associated breast cancer. Adv. Exp. Med. Biol.1252, 75–79 (2020). - PubMed
-
- Andersson, T. M. L., Johansson, A. L. V., Hsieh, C. C., Cnattingius, S. & Lambe, M. Increasing incidence of pregnancy-associated breast cancer in Sweden. Obstet. Gynecol.114, 568–572 (2009). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

