Exploration of phosphoproteomic association during epimorphic regeneration
- PMID: 39924536
- PMCID: PMC11808059
- DOI: 10.1038/s41598-024-84735-z
Exploration of phosphoproteomic association during epimorphic regeneration
Abstract
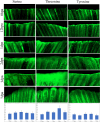
Unravelling the intricate patterns of site-specific protein phosphorylation during Epimorphic regeneration holds the key to unlocking the secrets of tissue complexity. Understanding these precise modifications and their impact on protein function could shed light on the remarkable regenerative capacity of tissues, with potential implications for therapeutic interventions. In this study we have systematically mapped the global phosphorylation modifications within regenerating tissue of zebrafish caudal fins, elucidating the intricate landscape of signalling pathway associate with the regeneration process. Based on mass spectrometry analysis, we identified 440 phosphorylated proteins using the immunoprecipitation method with phosphoserine, phosphothreonine, and phosphotyrosine antibodies, and 74 phosphorylated proteins using the TiO₂ column enrichment method were found differentially phosphorylated during the regeneration process from 12 hpa to 7 dpa compared to the control. Interestingly 95% of the proteins identified from TiO2 enrichment method were also found to be identified through the phosphoprotein antibody pull down method impacting the high accuracy and significance of the methods and greater association of the 70 proteins undergoing differential phosphorylation during the process of regeneration. Whole mount immunohistochemistry analysis reveals high association of phosphorylation at 1dpa, 2dpa and 3dpa regeneration time points. Network pathway analysis revealed that cancer-related diseases, organismal injuries and abnormalities as the most strongly associated canonical network pathways with the differentially expressed phosphoproteome in the mechanism of regeneration. This research enhances our comprehension on protein post-translational modification in the context of zebrafish caudal fin tissue regeneration, shedding light on its prospective application in the field of regenerative medicine.
Keywords: Immunoprecipitation; Phosphoprotein; Proteomics; Regeneration; TiO2; Zebrafish.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics approval: The experimental protocol was approved by Centre for Cellular and Molecular Biology institutional animal ethics committee (IAEC/CCMB/Protocol #50/2013).
Figures



References
-
- Akimenko, M. A., Marí-Beffa, M., Becerra, J. & Géraudie, J. Old questions, new tools, and some answers to the mystery of fin regeneration. Dev. Dyn.226 (2), 190–201 (2003). - PubMed
-
- Sehring, I. M. & Weidinger, G. Recent advancements in understanding fin regeneration in zebrafish. Wiley Interdiscip Rev. Dev. Biol.9(1), e367. (2020). - PubMed
-
- Banu, S. et al. Understanding the complexity of epimorphic regeneration in zebrafish caudal fin tissue: a transcriptomic and proteomic approach. Genomics114 (2), 110300 (2022). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

