The RNA M5C methyltransferase NSUN2 promotes progression of hepatocellular carcinoma by enhancing PKM2-mediated glycolysis
- PMID: 39924557
- PMCID: PMC11808121
- DOI: 10.1038/s41419-025-07414-5
The RNA M5C methyltransferase NSUN2 promotes progression of hepatocellular carcinoma by enhancing PKM2-mediated glycolysis
Abstract
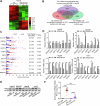
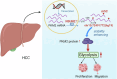
Hepatocellular carcinoma (HCC) is one of the most common cancers worldwide. The 5-methylcytosine (m5C) RNA methyltransferase NSUN2 is involved in cell proliferation and metastasis and is upregulated in a variety of cancers. However, the biological function and regulatory mechanism of NSUN2-mediated m5C modification have not been well studied in HCC. Our results showed that NSUN2 is upregulated and associated with poor prognosis in HCC patients after hepatectomy. NSUN2 overexpression significantly promoted HCC growth and metastasis, whereas NSUN2 knockdown had the opposite effect. m5C RNA immunoprecipitation sequencing (m5C-RIP-Seq) revealed that m5C hypermethylation correlates with mRNA overexpression and that NSUN2-mediated m5C hypermethylation promotes metabolism in HCC patients. Mechanistically, our data revealed that PKM2, a terminal enzyme in the glycolytic pathway, is a downstream target of NSUN2-mediated m5C modification. Specifically, NSUN2 could stabilize PKM2 mRNA by increasing the m5C level of the m5C site C773 in the 3'-UTR of PKM2 mRNA. In addition, rescue assays revealed that NSUN2 promotes HCC glycolysis and progression by upregulating PKM2. In conclusion, this study revealed that NSUN2-mediated m5C modification promotes glycolysis and the progression of hepatocellular carcinoma by stabilizing PKM2 mRNA, and provides a potential prognostic factor and therapeutic target for HCC patients.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: Human specimen collection was approved by the Ethics Committee of Eastern Hepatobiliary Surgery Hospital (approval number: EHBHKY2018-02-014). Written informed consent was obtained from each patient according to the policies of the committee. The animal experiments in this study conformed to the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines ( http://www.nc3rs.org.uk/arrive-guidelines ) and were approved by the Institutional Animal Care and Use Committee of Shanghai University of Traditional Chinese Medicine (Shanghai, China).
Figures





References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

