Radiogenomics of intrahepatic cholangiocarcinoma predicts immunochemotherapy response and identifies therapeutic target
- PMID: 39924997
- PMCID: PMC12260641
- DOI: 10.3350/cmh.2024.0895
Radiogenomics of intrahepatic cholangiocarcinoma predicts immunochemotherapy response and identifies therapeutic target
Abstract
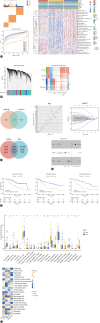
Background/aims: Identifying patients with intrahepatic cholangiocarcinoma (ICC) likely to benefit from immunochemotherapy, the new front-line treatment, remains challenging. We aimed to unveil a novel radiotranscriptomic signature that can facilitate treatment response prediction by multi-omics integration and multiscale modelling.
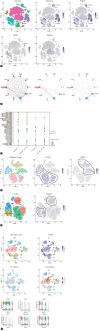
Methods: We analyzed bulk, single-cell and spatial transcriptomic data comprising 457 ICC patients to identify an immune-related score (IRS), followed by decoding its spatial immune context. We mapped radiomics profiles onto spatial-specific IRS using machine learning to define a novel radiotranscriptomic signature, followed by multi-scale and multi-cohort validation covering 331 ICC patients. The signature was further explored for the potential therapeutic target from in vitro to in vivo.
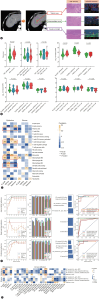
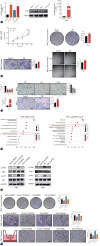
Results: We revealed a novel 3-gene (PLAUR, CD40LG, and FGFR4) IRS whose down-regulation correlated with better survival and improved sensitivity to immunochemotherapy. We highlighted functional IRS-immune interactions within tumor epithelium, rather than stromal compartment, irrespective of geospatial locations. Machine learning pipeline identified the optimal 3-feature radiotranscriptomic signature that was well-validated by immunohistochemical assays in molecular cohort, exhibited favorable external prognostic validity with C-index over 0.64 in resection cohort, and predicted treatment response with an area under the curve of up to 0.84 in immunochemotherapy cohort. We also showed that anti-uPAR/PLAUR alone or in combination with anti-programmed cell death protein 1 therapy remarkably curbed tumor growth, using in vitro ICC cell lines and in vivo humanized ICC patient-derived xenograft mouse models.
Conclusion: This proof-of-concept study sheds light on the spatially-resolved radiotranscriptomic signature to improve patient selection for emerging immunochemotherapy and high-order immunotherapy combinations in ICC.
Keywords: Intrahepatic cholangiocarcinoma; Machine learning; Multi-omics profiling; Prediction model; Radiogenomics.
Conflict of interest statement
The authors have no conflicts to disclose.
Figures




References
-
- Moris D, Palta M, Kim C, Allen PJ, Morse MA, Lidsky ME. Advances in the treatment of intrahepatic cholangiocarcinoma: An overview of the current and future therapeutic landscape for clinicians. CA Cancer J Clin. 2023;73:198–222. - PubMed
-
- EASL-ILCA Clinical Practice Guidelines on the management of intrahepatic cholangiocarcinoma. J Hepatol. 2023;79:181–208. - PubMed
-
- Kelley RK, Ueno M, Yoo C, Finn RS, Furuse J, Ren Z, et al. Pembrolizumab in combination with gemcitabine and cisplatin compared with gemcitabine and cisplatin alone for patients with advanced biliary tract cancer (KEYNOTE-966): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2023;401:1853–1865. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

