Pulmonary Aspergillosis and Low HIES Score in a Family with STAT3 N-Terminal Domain Mutation
- PMID: 39928202
- PMCID: PMC11811237
- DOI: 10.1007/s10875-025-01867-1
Pulmonary Aspergillosis and Low HIES Score in a Family with STAT3 N-Terminal Domain Mutation
Abstract
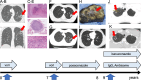
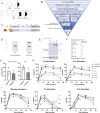
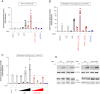
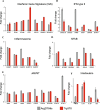
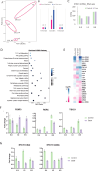
Signal transducer and activator of transcription 3 (STAT3) plays a key role in leukocytic and non-leukocytic cells. Germ line mutations in STAT3, which are mainly found in the SH2, DNA binding and transactivation domains, can be loss- or gain-of-function (LOF and GOF). STAT3 N-terminal domain (NTD) mutations are rare, and their biological effects remain incompletely understood. We explored the significance of STAT3 NTD p.Trp37* variant in a patient with chronic pulmonary aspergillosis and a low Hyper-IgE syndrome (HIES) score. In cell culture models, the expression of full-length p.Trp37* allele showed shorter STAT3 protein expression suggesting a re-initiation (Met99 or Met143). STAT3 activity using luciferase reporter assay showed a twofold-increased activity of the STAT3 p.Trp37* STAT3 protein compared with WT STAT3 at basal level and upon IL-6 stimulation. In contrast, the activity of the short pTrp37* peptide (amino acids 1 to 37) was amorphic but without dominant negative (DN) effect on transcriptional activity or STAT3 Tyr705 phosphorylation. The proteins initiated at Met99 and Met143 were surprisingly hypermorphic. In carriers' peripheral blood mononuclear cells (PBMCs), both WT and mutated STAT3 mRNA were equally present and the global amount of STAT3 protein was not significantly reduced. In stimulated heterozygous carriers' PBMCs, however, STAT3 Tyr705 phosphorylation and Th17 were reduced but not completely abolished. This suggests a DN effect of an unknown product of the p.Trp37* allele. Transcriptomics analysis of PBMCs from the index revealed selectively distinct gene expression. We conclude that heterozygosity for the NTD p.Trp37* STAT3 mutation defines a novel allelic form of STAT3 deficiency, associated with a chronic pulmonary aspergillosis and minor signs of HIES.
Keywords: Mutation; Pulmonary aspergillosis; STAT3.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval: The study was approved by Ethics Committee of Oulu University Hospital. Consent to Participate: Informed consent for participation was obtained from all individuals. Consent for Publication: Consent for publication was obtained from all individuals. Conflicts of Interest: The authors declare that they have no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

