Mitofusin 2 displays fusion-independent roles in proteostasis surveillance
- PMID: 39929801
- PMCID: PMC11811173
- DOI: 10.1038/s41467-025-56673-5
Mitofusin 2 displays fusion-independent roles in proteostasis surveillance
Abstract
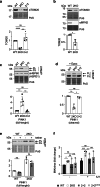
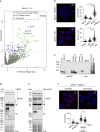
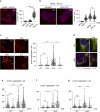
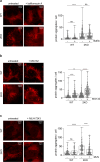
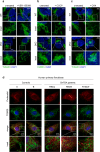
Mitochondria are essential organelles and their functional state dictates cellular proteostasis. However, little is known about the molecular gatekeepers involved, especially in absence of external stress. Here we identify a role of MFN2 in quality control independent of its function in organellar shape remodeling. MFN2 ablation alters the cellular proteome, marked for example by decreased levels of the import machinery and accumulation of the kinase PINK1. Moreover, MFN2 interacts with the proteasome and cytosolic chaperones, thereby preventing aggregation of newly translated proteins. Similarly to MFN2-KO cells, patient fibroblasts with MFN2-disease variants recapitulate excessive protein aggregation defects. Restoring MFN2 levels re-establishes proteostasis in MFN2-KO cells and rescues fusion defects of MFN1-KO cells. In contrast, MFN1 loss or mitochondrial shape alterations do not alter protein aggregation, consistent with a fusion-independent role of MFN2 in cellular homeostasis. In sum, our findings open new possibilities for therapeutic strategies by modulation of MFN2 levels.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Suomalainen, A. & Nunnari, J. Mitochondria at the crossroads of health and disease. Cell187, 2601–2627 (2024). - PubMed
-
- Giacomello, M., Pyakurel, A., Glytsou, C. & Scorrano, L. The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell Biol.21, 204–224 (2020). - PubMed
-
- D’Amico, D., Sorrentino, V. & Auwerx, J. Cytosolic proteostasis networks of the mitochondrial stress response. Trends Biochem Sci.42, 712–725 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

