Macrocycle-based PROTACs selectively degrade cyclophilin A and inhibit HIV-1 and HCV
- PMID: 39929804
- PMCID: PMC11811207
- DOI: 10.1038/s41467-025-56317-8
Macrocycle-based PROTACs selectively degrade cyclophilin A and inhibit HIV-1 and HCV
Abstract
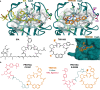
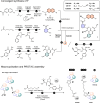
Targeting host proteins that are crucial for viral replication offers a promising antiviral strategy. We have designed and characterised antiviral PROteolysis TArgeting Chimeras (PROTACs) targeting the human protein cyclophilin A (CypA), a host cofactor for unrelated viruses including human immunodeficiency virus (HIV) and hepatitis C virus (HCV). The PROTAC warheads are based on fully synthetic macrocycles derived from sanglifehrin A, which are structurally different from the classical Cyp inhibitor, cyclosporine A. Our Cyp-PROTACs decrease CypA levels in cell lines and primary human cells and have high specificity for CypA confirmed by proteomics experiments. Critically, CypA degradation facilitates improved antiviral activity against HIV-1 in primary human CD4+ T cells compared to the non-PROTAC parental inhibitor, at limiting inhibitor concentrations. Similarly, we observe antiviral activity against HCV replicon in a hepatoma cell line. We propose that CypA-targeting PROTACs inhibit viral replication potently and anticipate reduced evolution of viral resistance and broad efficacy against unrelated viruses. Furthermore, they provide powerful tools for probing cyclophilin biology.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The Ciulli laboratory receives or has received sponsored research support from Almirall, Amgen, Amphista Therapeutics, Boehringer Ingelheim, Eisai, Merck KGaA, Nurix Therapeutics, Ono Pharmaceutical and Tocris-BioTechne. A.C. is a scientific founder, advisor, and shareholder of Amphista Therapeutics, a company that is developing targeted protein degradation therapeutic platforms. The other authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

