PAM-adjacent DNA flexibility tunes CRISPR-Cas12a off-target binding
- PMID: 39929897
- PMCID: PMC11811290
- DOI: 10.1038/s41598-025-87565-9
PAM-adjacent DNA flexibility tunes CRISPR-Cas12a off-target binding
Abstract
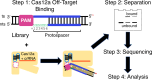
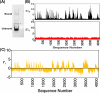
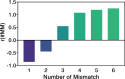
Cas12a is a class 2 type V CRISPR-associated nuclease that uses an effector complex comprised of a single protein activated by a CRISPR-encoded small RNA to cleave double-stranded DNA at specific sites. Cas12a processes unique features as compared to other CRISPR effector nucleases such as Cas9, and has been demonstrated as an effective tool for manipulating complex genomes. Prior studies have indicated that DNA flexibility at the region adjacent to the protospacer-adjacent-motif (PAM) contributes to Cas12a target recognition. Here, we adapted a SELEX-seq approach to further examine the connection between PAM-adjacent DNA flexibility and off-target binding by Cas12a. A DNA library containing DNA-DNA mismatches at PAM + 1 to + 6 positions was generated and subjected to binding in vitro with FnCas12a in the absence of pairing between the RNA guide and DNA target. The bound and unbound populations were sequenced to determine the propensity for off-target binding for each of the individual sequences. Analyzing the position and nucleotide dependency of the DNA-DNA mismatches showed that PAM-dependent Cas12a off-target binding requires unpairing of the protospacer at PAM + 1 and increases with unpairing at PAM + 2 and + 3. This revealed that PAM-adjacent DNA flexibility can tune Cas12a off-target binding. The work adds support to the notion that physical properties of the DNA modulate Cas12a target discrimination, and has implications on Cas12a-based applications.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.Mol Cell. 2017 Aug 17;67(4):633-645.e3. doi: 10.1016/j.molcel.2017.06.035. Epub 2017 Aug 3. Mol Cell. 2017. PMID: 28781234 Free PMC article.
-
Enhancement of target specificity of CRISPR-Cas12a by using a chimeric DNA-RNA guide.Nucleic Acids Res. 2020 Sep 4;48(15):8601-8616. doi: 10.1093/nar/gkaa605. Nucleic Acids Res. 2020. PMID: 32687187 Free PMC article.
-
Kinetic Basis for DNA Target Specificity of CRISPR-Cas12a.Mol Cell. 2018 Sep 6;71(5):816-824.e3. doi: 10.1016/j.molcel.2018.06.043. Epub 2018 Aug 2. Mol Cell. 2018. PMID: 30078724 Free PMC article.
-
Methods for decoding Cas9 protospacer adjacent motif (PAM) sequences: A brief overview.Methods. 2017 May 15;121-122:3-8. doi: 10.1016/j.ymeth.2017.03.006. Epub 2017 Mar 24. Methods. 2017. PMID: 28344037 Review.
-
CRISPR-Cas12a System for Biosensing and Gene Regulation.Chem Asian J. 2021 Apr 19;16(8):857-867. doi: 10.1002/asia.202100043. Epub 2021 Mar 18. Chem Asian J. 2021. PMID: 33638271 Review.
References
-
- Swarts, D. C. & Jinek, M. Cas9 versus Cas12a/Cpf1: structure-function comparisons and implications for genome editing. Wiley Interdiscip. Rev. RNA 9, e1481 (2018). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

