Mechanisms of NLRP3 activation and inhibition elucidated by functional analysis of disease-associated variants
- PMID: 39930093
- PMCID: PMC11876074
- DOI: 10.1038/s41590-025-02088-9
Mechanisms of NLRP3 activation and inhibition elucidated by functional analysis of disease-associated variants
Abstract
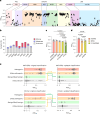
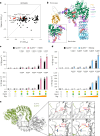
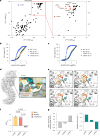
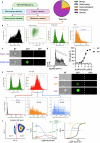
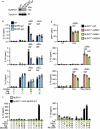
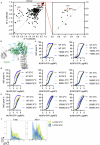
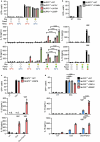
The NLRP3 inflammasome is a multiprotein complex that mediates caspase-1 activation and the release of proinflammatory cytokines, including interleukin (IL)-1β and IL-18. Gain-of-function variants in the gene encoding NLRP3 (also called cryopyrin) lead to constitutive inflammasome activation and excessive IL-1β production in cryopyrin-associated periodic syndromes (CAPS). Here we present functional screening and automated analysis of 534 NLRP3 variants from the international INFEVERS registry and the ClinVar database. This resource captures the effect of NLRP3 variants on ASC speck formation spontaneously, at low temperature, after inflammasome stimulation and with the specific NLRP3 inhibitor MCC950. Most notably, our analysis facilitated the updated classification of NLRP3 variants in INFEVERS. Structural analysis suggested multiple mechanisms by which CAPS variants activate NLRP3, including enhanced ATP binding, stabilizing the active NLRP3 conformation, destabilizing the inactive NLRP3 complex and promoting oligomerization of the pyrin domain. Furthermore, we identified pathogenic variants that can hypersensitize the activation of NLRP3 in response to nigericin and cold temperature exposure. We also found that most CAPS-related NLRP3 variants can be inhibited by MCC950; however, NLRP3 variants with changes to proline affecting helices near the inhibitor binding site are resistant to MCC950, as are variants in the pyrin domain, which likely trigger activation directly with the pyrin domain of ASC. Our findings could help stratify the CAPS population for NLRP3 inhibitor clinical trials and our automated methodologies can be implemented for molecules with a different mechanism of activation and in laboratories worldwide that are interested in adding new functionally validated NLRP3 variants to the resource. Overall, our study provides improved diagnosis for patients with CAPS, mechanistic insight into the activation of NLRP3 and stratification of patients for the future application of targeted therapeutics.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: S.L.M. is the Vice President of Discovery Biology of NRG Therapeutics and scientific advisor for Odyssey Therapeutics. M.G. is a scientific advisor to BioAge Labs. The other authors declare no competing interests.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

