Organoid modeling reveals the tumorigenic potential of the alveolar progenitor cell state
- PMID: 39930268
- PMCID: PMC11914084
- DOI: 10.1038/s44318-025-00376-6
Organoid modeling reveals the tumorigenic potential of the alveolar progenitor cell state
Abstract
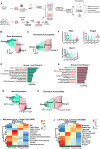
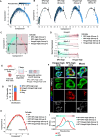
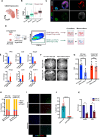
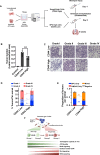
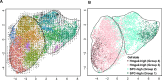
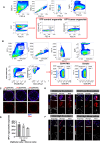
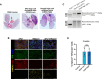
Cancers display cellular, genetic and epigenetic heterogeneity, complicating disease modeling. Multiple cell states defined by gene expression have been described in lung adenocarcinoma (LUAD). However, the functional contributions of cell state and the regulatory programs that control chromatin and gene expression in the early stages of tumor initiation are not well understood. Using single-cell RNA and ATAC sequencing in Kras/p53-driven tumor organoids, we identified two major cellular states: one more closely resembling alveolar type 2 (AT2) cells (SPC-high), and the other with epithelial-mesenchymal-transition (EMT)-associated gene expression (Hmga2-high). Each state exhibited distinct transcription factor networks, with SPC-high cells associated with TFs regulating AT2 fate and Hmga2-high cells enriched in Wnt- and NFκB-related TFs. CD44 was identified as a marker for the Hmga2-high state, enabling functional comparison of the two populations. Organoid assays and orthotopic transplantation revealed that SPC-high, CD44-negative cells exhibited higher tumorigenic potential within the lung microenvironment. These findings highlight the utility of organoids in understanding chromatin regulation in early tumorigenesis and identifying novel early-stage therapeutic targets in Kras-driven LUAD.
Keywords: Alveolar; Cell State; Kras; Lung Cancer; Organoids.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. We declare no competing interests. CFK and ALM are founders of Cellforma. CFK is a member of the EMBO Journal editorial advisory board.
Figures



Update of
-
Organoid modeling reveals the tumorigenic potential of the alveolar progenitor cell state.Res Sq [Preprint]. 2023 Mar 14:rs.3.rs-2663901. doi: 10.21203/rs.3.rs-2663901/v1. Res Sq. 2023. Update in: EMBO J. 2025 Mar;44(6):1804-1828. doi: 10.1038/s44318-025-00376-6. PMID: 36993454 Free PMC article. Updated. Preprint.
References
-
- Cassel TN, Nord M (2003) C/EBP transcription factors in the lung epithelium. Am J Physiol Lung Cell Mol Physiol 285:L773–L781 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

