MBNL overexpression rescues cardiac phenotypes in a myotonic dystrophy type 1 heart mouse model
- PMID: 39932794
- PMCID: PMC11957708
- DOI: 10.1172/JCI186416
MBNL overexpression rescues cardiac phenotypes in a myotonic dystrophy type 1 heart mouse model
Abstract
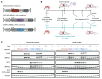
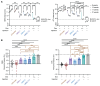
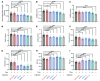
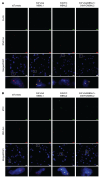
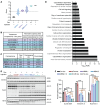
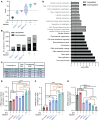
Myotonic dystrophy type 1 (DM1) is an autosomal dominant disease caused by a CTG repeat expansion in the dystrophia myotonica protein kinase (DMPK) gene. The expanded CUG repeat RNA (CUGexp RNA) transcribed from the mutant allele sequesters the muscleblind-like (MBNL) family of RNA-binding proteins, causing their loss of function and disrupting regulated pre-mRNA processing. We used a DM1 heart mouse model that inducibly expresses CUGexp RNA to test the contribution of MBNL loss to DM1 cardiac abnormalities and explored MBNL restoration as a potential therapy. AAV9-mediated overexpression of MBNL1 and/or MBNL2 significantly rescued DM1 cardiac phenotypes including conduction delays, contractile dysfunction, hypertrophy, and misregulated alternative splicing and gene expression. While robust, the rescue was partial compared with reduced CUGexp RNA and plateaued with increased exogenous MBNL expression. These findings demonstrate that MBNL loss is a major contributor to DM1 cardiac manifestations and suggest that additional mechanisms play a role, highlighting the complex nature of DM1 pathogenesis.
Keywords: Arrhythmias; Cardiovascular disease; Genetic diseases; Genetics; Therapeutics.
Conflict of interest statement
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

