TRIB3 mediates vascular calcification by facilitating self-ubiquitination and dissociation of Smurf1 in chronic kidney disease
- PMID: 39932798
- PMCID: PMC11957692
- DOI: 10.1172/JCI175972
TRIB3 mediates vascular calcification by facilitating self-ubiquitination and dissociation of Smurf1 in chronic kidney disease
Abstract
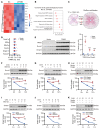
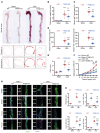
The osteogenic environment promotes vascular calcium phosphate deposition and aggregation of unfolded and misfolded proteins, resulting in ER stress in chronic kidney disease (CKD). Controlling ER stress through genetic intervention is a promising approach for treating vascular calcification. In this study, we demonstrated a positive correlation between ER stress-induced tribble homolog 3 (TRIB3) expression and progression of vascular calcification in human and rodent CKD. Increased TRIB3 expression promoted vascular smooth muscle cell (VSMC) calcification by interacting with the C2 domain of the E3 ubiquitin-protein ligase Smurf1, facilitating its K48-related self-ubiquitination at Lys381 and Lys383 and subsequent dissociation from the plasma membrane and nuclei. This degeneration of Smurf1 accelerated the stabilization of the osteogenic transcription factors RUNX family transcription factor 2 (Runx2) and SMAD family member 1 (Smad1). C/EBP homologous protein and activating transcription factor 4 are upstream transcription factors of TRIB3 in an osteogenic environment. Genetic KO of TRIB3 or rescue of Smurf1 ameliorated VSMC and vascular calcification by stabilizing Smurf1 and enhancing the degradation of Runx2 and Smad1. Our findings shed light on the vital role of TRIB3 as a scaffold in ER stress and vascular calcification and offer a potential therapeutic option for CKD.
Keywords: Cardiology; Cardiovascular disease; Ubiquitin-proteosome system; Vascular biology.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

