Concurrent Inhibition of the RAS-MAPK Pathway and PIKfyve Is a Therapeutic Strategy for Pancreatic Cancer
- PMID: 39932818
- PMCID: PMC11999774
- DOI: 10.1158/0008-5472.CAN-24-1757
Concurrent Inhibition of the RAS-MAPK Pathway and PIKfyve Is a Therapeutic Strategy for Pancreatic Cancer
Abstract
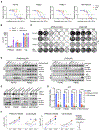
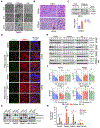
Pancreatic ductal adenocarcinoma (PDAC) is characterized by KRAS- and autophagy-dependent growth. Inhibition of the KRAS-RAF-MEK-ERK pathway enhances autophagic flux and dependency, and concurrent treatment with the nonspecific autophagy inhibitor chloroquine (CQ) and ERK-MAPK pathway inhibitors can synergistically block PDAC growth. However, CQ is limited in terms of specificity and potency. To find alternative anti-autophagy strategies, in this study, we performed a CRISPR-Cas9 loss-of-function screen in PDAC cell lines that identified the lipid kinase phosphatidylinositol-3-phosphate 5-kinase (PIKfyve) as a growth-promoting gene. PIKfyve inhibition by the small molecule apilimod resulted in durable growth suppression, with much greater potency than CQ treatment. PIKfyve inhibition caused lysosomal dysfunction, reduced autophagic flux, and led to the accumulation of autophagy-related proteins. Furthermore, PIKfyve inhibition blocked the compensatory increases in autophagic flux associated both with MEK inhibition and with direct RAS inhibition. Accordingly, combined inhibition of PIKfyve and the RAS-MAPK pathway showed robust growth suppression across a panel of KRAS-mutant PDAC models. Growth suppression was due, in part, to potentiated cell-cycle arrest and induction of apoptosis following loss of inhibitor of apoptosis proteins. These findings indicate that concurrent inhibition of RAS and PIKfyve is a synergistic, cytotoxic combination that may represent a therapeutic strategy for PDAC. Significance: PIKfyve inhibition effectively blocks autophagy in multiple models of KRAS-mutant pancreatic cancer and can synergize with inhibitors of members of the RAS-MAPK pathway, providing an effective combination strategy for pancreatic cancer.
©2025 American Association for Cancer Research.
Conflict of interest statement
K.L. Bryant has received research funding support from SpringWorks Therapeutics. A.D. Cox has consulted for Eli Lilly and Mirati Therapeutics. C.J. Der was a consultant/advisory board member for Cullgen, Deciphera Pharmaceuticals, Kestrel Therapeutics, Mirati Therapeutics, Reactive Biosciences, Revere Pharmaceuticals, Revolution Medicines, Ribometrix, Sanofi and SHY Therapeutics. C.J.D. has received research funding support from Deciphera Pharmaceuticals, Mirati Therapeutics, Reactive Biosciences, Revolution Medicines, and SpringWorks Therapeutics. E.F. Petricoin is a shareholder and consultant of Avant Diagnostics, TheraLink Technologies, Inc. and Perthera. E.F. Petricoin received funding support from Mirati Therapeutics, Genentech, Inc., and Abbvie, Inc.
Figures




References
MeSH terms
Substances
Grants and funding
- R01 CA262414/CA/NCI NIH HHS/United States
- T32GM135095/National Institute of General Medical Sciences (NIGMS)
- T32 CA009156/CA/NCI NIH HHS/United States
- F32CA232529/National Cancer Institute (NCI)
- P30 CA016086/CA/NCI NIH HHS/United States
- F32 CA232529/CA/NCI NIH HHS/United States
- 22-WG-DERB/Pancreatic Cancer Action Network (PCAN)
- U01 CA199235/CA/NCI NIH HHS/United States
- T32CA071341/National Cancer Institute (NCI)
- Pancreatic Cancer Action Network (PCAN)
- T32 CA071341/CA/NCI NIH HHS/United States
- 388222/Lustgarten Foundation (Lustgarten)
- National Pancreas Foundation (NPF)
- P50 CA196510/CA/NCI NIH HHS/United States
- W81XWH2110693/U.S. Department of Defense (DOD)
- 15-70-25-BRYA/Pancreatic Cancer Action Network (PCAN)
- Sky Foundation (THE SKY FOUNDATION)
- T32 GM135095/GM/NIGMS NIH HHS/United States
- T32 CA244125/CA/NCI NIH HHS/United States
- P50CA196510/National Cancer Institute (NCI)
- P50 CA257911/CA/NCI NIH HHS/United States
- R01CA42978/National Cancer Institute (NCI)
- PF-22-066-01-TBE/American Cancer Society (ACS)
- 15-70-25-BRYA/American Association for Cancer Research (AACR)
- R37 CA251877/CA/NCI NIH HHS/United States
- R01 CA042978/CA/NCI NIH HHS/United States
- P01 CA203657/CA/NCI NIH HHS/United States
- R35 CA232113/CA/NCI NIH HHS/United States
- F31 CA284869/CA/NCI NIH HHS/United States
- R37CA251877/National Cancer Institute (NCI)
- T32CA009156/National Cancer Institute (NCI)
- W81XWH2110692/U.S. Department of Defense (DOD)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

