A guanidine-based coronavirus replication inhibitor which targets the nsp15 endoribonuclease and selects for interferon-susceptible mutant viruses
- PMID: 39932973
- PMCID: PMC11856660
- DOI: 10.1371/journal.ppat.1012571
A guanidine-based coronavirus replication inhibitor which targets the nsp15 endoribonuclease and selects for interferon-susceptible mutant viruses
Abstract
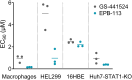
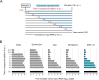
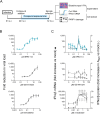
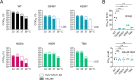
The approval of COVID-19 vaccines and antiviral drugs has been crucial to end the global health crisis caused by SARS-CoV-2. However, to prepare for future outbreaks from drug-resistant variants and novel zoonotic coronaviruses (CoVs), additional therapeutics with a distinct antiviral mechanism are needed. Here, we report a novel guanidine-substituted diphenylurea compound that suppresses CoV replication by interfering with the uridine-specific endoribonuclease (EndoU) activity of the viral non-structural protein-15 (nsp15). This compound, designated EPB-113, exhibits strong and selective cell culture activity against human coronavirus 229E (HCoV-229E) and also suppresses the replication of SARS-CoV-2. Viruses, selected under EPB-113 pressure, carried resistance sites at or near the catalytic His250 residue of the nsp15-EndoU domain. Although the best-known function of EndoU is to avoid induction of type I interferon (IFN-I) by lowering the levels of viral dsRNA, EPB-113 was found to mainly act via an IFN-independent mechanism, situated during viral RNA synthesis. Using a combination of biophysical and enzymatic assays with the recombinant nsp15 proteins from HCoV-229E and SARS-CoV-2, we discovered that EPB-113 enhances the EndoU cleavage activity of hexameric nsp15, while reducing its thermal stability. This mechanism explains why the virus escapes EPB-113 by acquiring catalytic site mutations which impair compound binding to nsp15 and abolish the EndoU activity. Since the EPB-113-resistant mutant viruses induce high levels of IFN-I and its effectors, they proved unable to replicate in human macrophages and were readily outcompeted by the wild-type virus upon co-infection of human fibroblast cells. Our findings suggest that antiviral targeting of nsp15 can be achieved with a molecule that induces a conformational change in this protein, resulting in higher EndoU activity and impairment of viral RNA synthesis. Based on the appealing mechanism and resistance profile of EPB-113, we conclude that nsp15 is a challenging but highly relevant drug target.
Copyright: © 2025 Van Loy et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- World Health Organization. WHO COVID-19 dashboard. [cited 2025 Jan 08]. Available from: https://data.who.int/dashboards/covid19/cases.
-
- Infectious Diseases Society of America. IDSA guidelines on the treatment and management of patients with COVID-19. [cited 2024 Aug 23]. Available from: https://www.idsociety.org/COVID19guidelines#null.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

