Unraveling AURKB as a potential therapeutic target in pulmonary hypertension using integrated transcriptomic analysis and pre-clinical studies
- PMID: 39933527
- PMCID: PMC11866512
- DOI: 10.1016/j.xcrm.2025.101964
Unraveling AURKB as a potential therapeutic target in pulmonary hypertension using integrated transcriptomic analysis and pre-clinical studies
Abstract
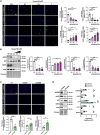
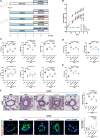
Despite advances in treatment, the prognosis for patients with pulmonary arterial hypertension (PAH) remains dismal, highlighting the need for further therapeutic advances. By using RNA sequencing on pulmonary artery smooth muscle cells (PASMCs), functional enrichment, and connectivity map analyses, we identify Aurora kinase B (AURKB) as a candidate therapeutic target. We show that AURKB inhibition blocks cell cycle progression and reverses the gene signature of PAH-PASMCs. We also report that PAH-PASMCs that escape apoptosis acquire a senescence-associated secretory phenotype. In vivo, AURKB inhibition using barasertib improves hemodynamics in two preclinical models of established PAH by attenuating pulmonary vascular remodeling. A therapeutic effect is also observed in human precision-cut lung slices. Finally, we demonstrate that the combination of barasertib with a p21 attenuator is more effective in reducing vascular remodeling than either drug alone. These findings provide insight into strategies for therapeutic manipulation.
Keywords: FOXM1; aurora kinase B; mitosis; pulmonary arterial hypertension; right ventricular failure; senescence; vascular remodeling.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures






References
-
- Hoeper M.M., Badesch D.B., Ghofrani H.A., Gibbs J.S.R., Gomberg-Maitland M., McLaughlin V.V., Preston I.R., Souza R., Waxman A.B., Grünig E., et al. Phase 3 Trial of Sotatercept for Treatment of Pulmonary Arterial Hypertension. N. Engl. J. Med. 2023;388:1478–1490. doi: 10.1056/NEJMoa2213558. - DOI - PubMed
-
- Pullamsetti S.S., Savai R., Seeger W., Goncharova E.A. Translational Advances in the Field of Pulmonary Hypertension. From Cancer Biology to New Pulmonary Arterial Hypertension Therapeutics. Targeting Cell Growth and Proliferation Signaling Hubs. Am. J. Respir. Crit. Care Med. 2017;195:425–437. doi: 10.1164/rccm.201606-1226PP. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

