The spatiotemporal transcriptional profiling of murine brain during cerebral malaria progression and after artemisinin treatment
- PMID: 39934099
- PMCID: PMC11814382
- DOI: 10.1038/s41467-024-52223-7
The spatiotemporal transcriptional profiling of murine brain during cerebral malaria progression and after artemisinin treatment
Abstract
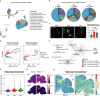
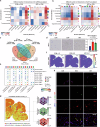
Cerebral malaria (CM) is a severe encephalopathy caused by Plasmodium parasite infection, resulting in thousands of annual deaths and neuro-cognitive sequelae even after anti-malarial drugs treatment. Despite efforts to dissect the mechanism, the cellular transcriptomic reprogramming within the spatial context remains elusive. Here, we constructed single-cell and spatial transcriptome atlases of experimental CM (ECM) male murine brain tissues with or without artesunate (ART) treatment. We identified activated inflammatory endothelial cells during ECM, characterized by a disrupted blood-brain barrier, increased antigen presentation, and leukocyte adhesion. We also observed that inflammatory microglia enhance antigen presentation pathway such as MHC-I to CD8+ cytotoxic T cells. The latter underwent an inflammatory state transition with up-regulated cytokine expression and cytotoxic activity. Multi-omics analysis revealed that the activated interferon-gamma response of injured neurons during ECM and persisted after ART treatment. Overall, our research provides valuable resources for understanding malaria parasite-host interaction mechanisms and adjuvant therapy development.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

