BBOX1 restrains TBK1-mTORC1 oncogenic signaling in clear cell renal cell carcinoma
- PMID: 39934163
- PMCID: PMC11814379
- DOI: 10.1038/s41467-025-56955-y
BBOX1 restrains TBK1-mTORC1 oncogenic signaling in clear cell renal cell carcinoma
Abstract
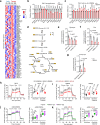
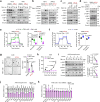
Clear cell renal cell carcinoma (ccRCC), a metabolic disease originating from renal proximal convoluted tubule (PCT) epithelial cells, remains incompletely understood in terms of its initiating signaling events. Here, we identify γ-butyrobetaine hydroxylase 1 (BBOX1), a key enzyme in carnitine synthesis predominantly expressed in PCT cells, as a tumor suppressor in ccRCC. BBOX1 expression is lost during ccRCC malignant transformation, and its restoration reduces cell viability in physiological medium and inhibits xenograft tumor growth. Transcriptomic analyses reveal that BBOX1 suppresses critical metabolic pathways including mTORC1 signaling and glycolysis in ccRCC. Further, we identify TANK-binding kinase 1 (TBK1) as an essential mediator of mTORC1 and glycolysis activation and as a target of BBOX1-mediated tumor suppression. Mechanistically, BBOX1 disrupts TBK1 activation by preventing its interaction with the upstream activator doublecortin-like kinase 2 (DCLK2). This BBOX1-DCLK2-TBK1 axis unveils an important mechanism in ccRCC metabolic dysregulation and highlights potential therapeutic strategies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Q.Z. received the consultation fee from Exelixis. Other authors declare no competing interests.
Figures





References
-
- Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin.72, 7–33 (2022). - PubMed
-
- Godwin, J. L., Zibelman, M., Plimack, E. R. & Geynisman, D. M. Immune checkpoint blockade as a novel immunotherapeutic strategy for renal cell carcinoma: a review of clinical trials. Discov. Med.18, 341–350 (2014). - PubMed
-
- Kaelin, W. G. Jr. Molecular basis of the VHL hereditary cancer syndrome. Nat. Rev. Cancer2, 673–682 (2002). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

