Mitochondrial DNA removal is essential for sperm development and activity
- PMID: 39934414
- PMCID: PMC11914152
- DOI: 10.1038/s44318-025-00377-5
Mitochondrial DNA removal is essential for sperm development and activity
Abstract
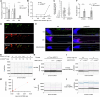
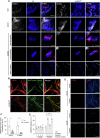
Active mitochondrial DNA (mtDNA) elimination during spermatogenesis has emerged as a conserved mechanism ensuring the uniparental mitochondrial inheritance in animals. However, given the existence of post-fertilization processes degrading sperm mitochondria, the physiological significance of mtDNA removal during spermatogenesis is not clear. Here we show that mtDNA clearance is indispensable for sperm development and activity. We uncover a previously unappreciated role of Poldip2 as a mitochondrial exonuclease that is specifically expressed in late spermatogenesis and required for sperm mtDNA elimination in Drosophila. Loss of Poldip2 impairs mtDNA clearance in elongated spermatids and impedes the progression of individualization complexes that strip away cytoplasmic materials and organelles. Over time, poldip2 mutant sperm exhibit marked nuclear genome fragmentation, and the flies become completely sterile. Notably, these phenotypes were rescued by expressing a mitochondrially targeted bacterial exonuclease, which ectopically removes mtDNA. Our work illustrates the developmental necessity of mtDNA clearance for effective cytoplasm removal at the end of spermatid morphogenesis, and for preventing potential nuclear-mitochondrial genome imbalance in mature sperm, in which nuclear genome activity is shut down.
Keywords: Drosophila spermatogenesis; EndoG; Exonuclease; Male Sterile; Maternal Inheritance.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures








References
-
- Bogenhagen DF, Rousseau D, Burke S (2008) The layered structure of human mitochondrial DNA nucleoids. J Biol Chem 283:3665–3675 - PubMed
-
- Chen D, McKearin DM (2003) A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development 130:1159–1170 - PubMed
-
- Chen Z, Zhang F, Xu H (2019) Human mitochondrial DNA diseases and Drosophila models. J Genet Genomics 46:201–212 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

