Small molecule treatment alleviates photoreceptor cilia defects in LCA5-deficient human retinal organoids
- PMID: 39934925
- PMCID: PMC11817871
- DOI: 10.1186/s40478-025-01943-y
Small molecule treatment alleviates photoreceptor cilia defects in LCA5-deficient human retinal organoids
Erratum in
-
Correction: Small molecule treatment alleviates photoreceptor cilia defects in LCA5-deficient human retinal organoids.Acta Neuropathol Commun. 2025 May 19;13(1):104. doi: 10.1186/s40478-025-02010-2. Acta Neuropathol Commun. 2025. PMID: 40390152 Free PMC article. No abstract available.
Abstract
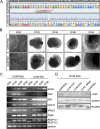
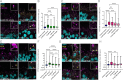
Bialleleic pathogenic variants in LCA5 cause one of the most severe forms of Leber congenital amaurosis, an early-onset retinal disease that results in severe visual impairment. Here, we report the use of gene editing to generate isogenic LCA5 knock-out (LCA5 KO) induced pluripotent stem cells (iPSC) and their differentiation to retinal organoids. The molecular and cellular phenotype of the LCA5 KO retinal organoids was studied in detail and compared to isogenic controls as well as patient-derived retinal organoids. The absence of LCA5 was confirmed in retinal organoids by immunohistochemistry and western blotting. There were no major changes in retinal organoid differentiation or ciliation, however, the localisation of CEP290 and IFT88 was significantly altered in LCA5 KO and patient photoreceptor cilia with extension along the axoneme. The LCA5-deficient organoids also had shorter outer segments and rhodopsin was mislocalised to the outer nuclear layer. We also identified transcriptomic and proteomic changes associated with the loss of LCA5. Importantly, treatment with the small molecules eupatilin, fasudil or a combination of both drugs reduced CEP290 and IFT88 accumulation along the cilia. The treatments also improved rhodopsin traffic to the outer segment and reduced mislocalisation of rhodopsin in the outer nuclear layer (ONL). The improvements in cilia-associated protein localisation and traffic were accompanied by significant changes in the transcriptome towards control gene expression levels in many of the differentially expressed genes. In summary, iPSC-derived retinal organoids are a powerful model for investigating the molecular and cellular changes associated with loss of LCA5 function and highlight the therapeutic potential of small molecules to treat retinal ciliopathies.
Keywords: Cilia; Gene editing; LCA; LCA5; Organoid; Photoreceptor; Retina; Retinal dystrophy; Stem cell; Therapy.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The control and JB342 patient iPSC lines used in this study are established lines that have been published previously and were obtained with appropriate consent and ethical approval. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





References
-
- den Hollander AI, Roepman R, Koenekoop RK, Cremers FPM (2008) Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog Retin Eye Res 27:391–419. 10.1016/j.preteyeres.2008.05.003 - PubMed
-
- Kumaran N, Pennesi ME, Yang P, Trzupek KM, Schlechter C, Moore AT, Weleber RG, Michaelides M (1993) Leber Congenital Amaurosis / early-onset severe retinal dystrophy overview. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A (eds) GeneReviews®. University of Washington, Seattle, Seattle (WA) - PubMed
-
- Kondkar AA, Abu-Amero KK (2019) Leber congenital amaurosis: current genetic basis, scope for genetic testing and personalized medicine. Exp Eye Res 189:107834. 10.1016/j.exer.2019.107834 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

