Identification of WDR74 and TNFRSF12A as biomarkers for early osteoarthritis using machine learning and immunohistochemistry
- PMID: 39935469
- PMCID: PMC11810735
- DOI: 10.3389/fimmu.2025.1517646
Identification of WDR74 and TNFRSF12A as biomarkers for early osteoarthritis using machine learning and immunohistochemistry
Abstract
Background: Osteoarthritis (OA) is a chronic joint condition that causes pain, limited mobility, and reduced quality of life, posing a threat to healthy aging. Early detection is crucial for improving prognosis. Recent research has focused on the role of ubiquitination-related genes (URGs) in early OA prediction. This study aims to integrate URG expression data with machine learning (ML) to identify biomarkers that improve diagnosis and prognosis in the early stages of OA.
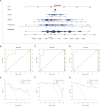
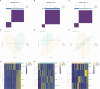
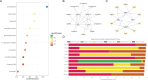
Methods: OA single-cell RNA sequencing datasets were collected from the GEO database. Single-cell analysis was performed to investigate the composition and relationships of chondrocytes in OA. The potential intercellular communication mechanisms were explored using the CellChat R package. URGs were retrieved from GeneCards, and ubiquitination scores were calculated using the AUCell package. Gene module analysis based on co-expression network analysis was conducted to identify core genes. Additionally, ML analysis was performed to identify core URGs and construct a diagnostic model. We employed XGBoost, a gradient-boosting ML algorithm, to identify core URGs and construct a diagnostic model. The model's performance was evaluated using the area under the curve (AUC) of the receiver operating characteristic (ROC) curve. In addition, we explored the relationship between core URGs and immune processes. The ChEA3 database was utilized to predict the transcription factors regulated by core ubiquitination-related genes. The expression of select URGs was validated using qRT-PCR and immunohistochemistry (IHC).
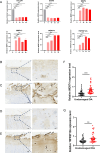
Results: We identified WDR74 and TNFRSF12A as pivotal ubiquitination-related genes associated with OA, exhibiting considerable differential expression. The diagnostic model constructed with URGs exhibited remarkable accuracy, with area under the curve (AUC) values consistently exceeding 0.9. The expression levels of WDR74 and TNFRSF12A were significantly higher in the IL-1β-induced group in an in vitro qPCR experiment. The IHC validation on human knee joint specimens confirmed the upregulation of WDR74 and TNFRSF12A in OA tissues, corroborating their potential as diagnostic biomarkers.
Conclusions: WDR74 and TNFRSF12A as principal biomarkers highlighted their attractiveness as therapeutic targets. The identification of core biomarkers might facilitate early intervention options, potentially modifying the illness trajectory and enhancing patient outcomes.
Keywords: diagnosis; machine learning; osteoarthritis; single-cell RNA sequencing; ubiquitination.
Copyright © 2025 Chen, Lin, Shi, Miao, Xue, Liu, Wang and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Peat G, Thomas M. Osteoarthritis year in review 2020: epidemiology & therapy. Osteoarthritis and Cartillage. (2021), 180–9. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

