The prevalence and antimicrobial resistance of respiratory pathogens isolated from feedlot cattle in Canada
- PMID: 39935643
- PMCID: PMC11810952
- DOI: 10.3389/fmicb.2025.1497402
The prevalence and antimicrobial resistance of respiratory pathogens isolated from feedlot cattle in Canada
Abstract
Objectives: The purpose of this study was to characterize the prevalence of antimicrobial resistance in Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni isolated from healthy feedlot cattle over 2 years, and investigate factors potentially associated with recovery of resistant isolates.
Methods: Deep-guarded nasopharyngeal (NP) swabs were used to sample feedlot cattle in multiple randomly selected feedlots (2019 n = 21, 2020 n = 26) at 2 timepoints. NP swabs were collected from 16 animals in each enrolled group upon entry processing and later in the feeding period. Cattle from the same groups (not necessarily the same animals) were sampled at both timepoints. Susceptibility testing was performed using the broth microdilution.
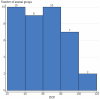
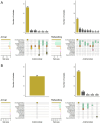
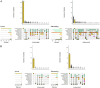
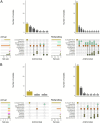
Results: A total of 1,392 cattle within 47 housing groups were sampled over 2 years, providing 625 bacterial isolates for investigation. Pasteurella multocida (27.4%) was the most frequently isolated BRD organism, followed by H. somni (9%) and M. haemolytica (8.5%). Resistance to ≥3 antimicrobial classes was detected in 2.4% of M. haemolytica, 3.4% of H. somni, and 21.3% of P. multocida isolates. Potential associations were investigated between recovery of resistant organisms and time of year at sampling (quarter), sampling timepoint (arrival or second sample), days on feed (DOF) at sampling, animal age categories, and BRD risk categories. There was a significant (p < 0.05) increase in resistance prevalence after arrival for macrolide drugs in M. haemolytica, and for ampicillin, danofloxacin, enrofloxacin, spectinomycin, gamithromycin, tildipirosin, tulathromycin and tetracycline in P. multocida isolates. Resistance was higher in calves than in yearlings for tulathromycin in H. somni, and for gamithromycin, spectinomycin, tulathromycin, tildipirosin, and tetracycline for P. multocida (p < 0.05) Resistance to tetracycline, tildipirosin, and tulathromycin decreased between 61-80 DOF and 81-100 DOF when compared to 20-40 DOF, whereas for spectinomycin, resistance was lower in cattle sampled between 61-80 DOF than those sampled at 20-40 DOF for P. multocida.
Discussion: The diversity of AMR profiles and associated risk factors between the BRD pathogens studied, underscores the importance of including all three organisms in future AMR studies in beef cattle.
Keywords: Histophilus somni; Mannheimia haemolytica; Pasteurella multocida; antimicrobial resistance; bovine respiratory disease.
Copyright © 2025 Rattanapanadda, Ramsay, Butters, Booker, Hannon, Hendrick, Van Donkersgoed, Warr, Gow and Morley.
Conflict of interest statement
JD was employed by Dr. Joyce Van Donkersgoed Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Abi Younes J., Ramsay D. E., Lacoste S., Deschner D., Hill J. E., Campbell J., et al. . (2022). Changes in the phenotypic susceptibility of Mannheimia haemolytica isolates to macrolide antimicrobials during the early feeding period following metaphylactic tulathromycin use in western Canadian feedlot calves. Can. Vet. J. 63, 920–928. - PMC - PubMed
-
- Andres-Lasheras S., Ha R., Zaheer R., Lee C., Booker C. W., Dorin C., et al. . (2021). Prevalence and risk factors associated with antimicrobial resistance in bacteria related to bovine respiratory disease-a broad cross-sectional study of beef cattle at entry into Canadian feedlots. Front. Vet. Sci. 8:692646. doi: 10.3389/fvets.2021.692646, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

