This is a preprint.
Genome-wide absolute quantification of chromatin looping
- PMID: 39935886
- PMCID: PMC11812599
- DOI: 10.1101/2025.01.13.632736
Genome-wide absolute quantification of chromatin looping
Abstract
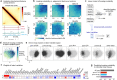
3D genomics methods such as Hi-C and Micro-C have uncovered chromatin loops across the genome and linked these loops to gene regulation. However, these methods only measure 3D interaction probabilities on a relative scale. Here, we overcome this limitation by using live imaging data to calibrate Micro-C in mouse embryonic stem cells, thus obtaining absolute looping probabilities for 36,804 chromatin loops across the genome. We find that the looped state is generally rare, with a mean probability of 2.3% and a maximum of 26% across the quantified loops. On average, CTCF-CTCF loops are stronger than loops between cis-regulatory elements (3.2% vs. 1.1%). Our findings can be extended to human stem cells and differentiated cells under certain assumptions. Overall, we establish an approach for genome-wide absolute loop quantification and report that loops generally occur with low probabilities, generalizing recent live imaging results to the whole genome.
Conflict of interest statement
Competing Interests The authors declare no competing interests.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
