Maize transcriptome profiling reveals low temperatures affect photosynthesis during the emergence stage
- PMID: 39935955
- PMCID: PMC11810925
- DOI: 10.3389/fpls.2025.1527447
Maize transcriptome profiling reveals low temperatures affect photosynthesis during the emergence stage
Abstract
Introduction: Earlier sowing is a promising strategy of ensuring sufficiently high maize yields in the face of negative environmental factors caused by climate change. However, it leads to the low temperature exposure of maize plants during emergence, warranting a better understanding of their response and acclimation to suboptimal temperatures.
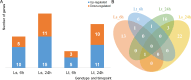
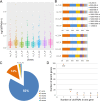
Materials and methods: To achieve this goal, whole transcriptome sequencing was performed on two maize inbred lines - tolerant/susceptible to low temperatures, at the 5-day-old seedling stage. Sampling was performed after 6h and 24h of treatment (10/8°C). The data was filtered, mapped, and the identified mRNAs, lncRNAs, and circRNAs were quantified. Expression patterns of the RNAs, as well as the interactions between them, were analyzed to reveal the ones important for low-temperature response.
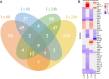
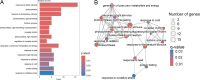
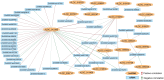
Results and discussion: Genes involved in different steps of photosynthesis were downregulated in both genotypes: psa, psb, lhc, and cab genes important for photosystem I and II functioning, as well as rca, prk, rbcx1 genes necessary for the Calvin cycle. The difference in low-temperature tolerance between genotypes appeared to arise from their ability to mitigate damage caused by photoinhibition: ctpa2, grx, elip, UF3GT genes showed higher expression in the tolerant genotype. Certain identified lncRNAs also targeted these genes, creating an interaction network induced by the treatment (XLOC_016169-rca; XLOC_002167-XLOC_006091-elip2). These findings shed light on the potential mechanisms of low-temperature acclimation during emergence and lay the groundwork for subsequent analyses across diverse maize genotypes and developmental stages. As such, it offers valuable guidance for future research directions in the molecular breeding of low-temperature tolerant maize.
Keywords: climate change; low temperature stress; maize; photosynthesis; whole transcriptome profiling.
Copyright © 2025 Božić, Ignjatović Micić, Anđelković, Delić and Nikolić.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Andrews S. 2010. Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed March 14, 2024).
-
- Balassa G., Oláh C., Balassa K., Rácz I., Kátay G., Kalapos B., et al. . (2022). Physiological and molecular background of maize cold-tolerance enhancement with S-methylmethionine salicylate. J. Plant Growth Regul. 41, 2073–2091. doi: 10.1007/s00344-022-10695-1 - DOI
-
- Banović Đeri B., Božić M., Dudić D., Vićić I., Milivojević M., Ignjatović-Micić D., et al. . (2022). Leaf transcriptome analysis of Lancaster versus other heterotic groups’ maize inbred lines revealed different regulation of cold-responsive genes. J. Agron. Crop Sci. 208, 497–509. doi: 10.1111/jac.12529 - DOI
-
- Bassu S., Fumagalli D., Toreti A., Ceglar A., Giunta F., Motzo R., et al. . (2021). Modelling potential maize yield with climate and crop conditions around flowering. Field Crops Res. 271, 108226. doi: 10.1016/j.fcr.2021.108226 - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

