Transcriptional repression of autophagy and lysosome biogenesis
- PMID: 39936623
- PMCID: PMC12087643
- DOI: 10.1080/15548627.2025.2465404
Transcriptional repression of autophagy and lysosome biogenesis
Abstract
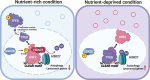
The microphthalmia/transcription factor E (MiT/TFE) family activates macroautophagy/autophagy and lysosomal genes during acute nutrient deficiency. However, the mechanisms that suppress transcription of these genes under steady-state, nutrient-rich conditions to prevent unnecessary expression remain unclear. In this study, we identified a previously unrecognized mechanism of transcriptional repression for autophagy and lysosomal genes. Under nutrient-rich conditions, USF2 (upstream transcription factor 2) binds to the coordinated lysosomal expression and regulation (CLEAR) motif, recruiting a repressive complex containing HDAC (histone deacetylase). In contrast, during nutrient deficiency, TFEB (transcription factor EB) displaces USF2 at the same motif, activating transcription. This switch is regulated by USF2 phosphorylation at serine 155 by GSK3B (glycogen synthase kinase 3 beta). Reduced phosphorylation under nutrient-deprived conditions weakens USF2's DNA binding affinity, allowing TFEB to competitively bind and activate target genes. Knockdown or knockout of Usf2 upregulates specific autophagy and lysosomal genes, leading to enhanced lysosomal functionality and increased autophagic flux. In USF2-deficient cells, the SERPINA1 Z variant/antitrypsin Z - an aggregation-prone mutant protein used as a model - is rapidly cleared via the autophagy-lysosome pathway. Therefore, modulation of USF2 activity may be a therapeutic strategy for managing diseases associated with autophagy and lysosomal dysfunction.Abbreviation: CLEAR: coordinated lysosomal expression and regulation; GSK3B: glycogen synthase kinase 3 beta; HDAC: histone deacetylase; MiT/TFE: microphthalmia/transcription factor E; NuRD: nucleosome remodeling and deacetylation; SERPINA1 Z variant/ATZ/antitrypsin Z; TFE3: transcription factor E3; TFEB: transcription factor EB; USF2: upstream transcription factor 2.
Keywords: Autophagy; MiT/TFE; TFEB; USF2; lysosome; transcriptional repressor.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
Similar articles
-
USF2 and TFEB compete in regulating lysosomal and autophagy genes.Nat Commun. 2024 Sep 27;15(1):8334. doi: 10.1038/s41467-024-52600-2. Nat Commun. 2024. PMID: 39333072 Free PMC article.
-
Induction of lysosome biogenesis is a novel function of the CGAS-STING1 pathway.Autophagy. 2025 May;21(5):1163-1164. doi: 10.1080/15548627.2025.2456064. Epub 2025 Jan 27. Autophagy. 2025. PMID: 39835593
-
Buddleoside alleviates nonalcoholic steatohepatitis by targeting the AMPK-TFEB signaling pathway.Autophagy. 2025 Jun;21(6):1316-1334. doi: 10.1080/15548627.2025.2466145. Epub 2025 Mar 16. Autophagy. 2025. PMID: 39936600
-
The Spectrum of Renal "TFEopathies": Flipping the mTOR Switch in Renal Tumorigenesis.Physiology (Bethesda). 2024 Nov 1;39(6):0. doi: 10.1152/physiol.00026.2024. Epub 2024 Jul 16. Physiology (Bethesda). 2024. PMID: 39012319 Review.
-
The complex relationship between TFEB transcription factor phosphorylation and subcellular localization.EMBO J. 2018 Jun 1;37(11):e98804. doi: 10.15252/embj.201798804. Epub 2018 May 15. EMBO J. 2018. PMID: 29764979 Free PMC article. Review.
Cited by
-
A biomimetic multimodal nanoplatform combining neutrophil-coated two-dimensional metalloporphyrinic framework nanosheet and exendin-4 to treat obesity-related osteoporosis.Mater Today Bio. 2025 Jun 21;33:102009. doi: 10.1016/j.mtbio.2025.102009. eCollection 2025 Aug. Mater Today Bio. 2025. PMID: 40673132 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
