CASP8 intronic expansion identified by poly-glycine-arginine pathology increases Alzheimer's disease risk
- PMID: 39937857
- PMCID: PMC11848317
- DOI: 10.1073/pnas.2416885122
CASP8 intronic expansion identified by poly-glycine-arginine pathology increases Alzheimer's disease risk
Abstract
Alzheimer's disease (AD) affects more than 10% of the population ≥65 y of age, but the underlying biological risks of most AD cases are unclear. We show anti-poly-glycine-arginine (a-polyGR) positive aggregates frequently accumulate in sporadic AD autopsy brains (45/80 cases). We hypothesize that these aggregates are caused by one or more polyGR-encoding repeat expansion mutations. We developed a CRISPR/deactivated-Cas9 enrichment strategy to identify candidate GR-encoding repeat expansion mutations directly from genomic DNA isolated from a-polyGR(+) AD cases. Using this approach, we isolated an interrupted (GGGAGA)n intronic expansion within a SINE-VNTR-Alu element in CASP8 (CASP8-GGGAGAEXP). Immunostaining using a-polyGR and locus-specific C-terminal antibodies demonstrate that the CASP8-GGGAGAEXP expresses hybrid poly(GR)n(GE)n(RE)n proteins that accumulate in CASP8-GGGAGAEXP(+) AD brains. In cells, expression of CASP8-GGGAGAEXP minigenes leads to increased p-Tau (Ser202/Thr205) levels. Consistent with other types of repeat-associated non-AUG (RAN) proteins, poly(GR)n(GE)n(RE)n protein levels are increased by stress. Additionally, levels of these stress-induced proteins are reduced by metformin. Association studies show specific aggregate promoting interrupted CASP8-GGGAGAEXP sequence variants found in ~3.6% of controls and 7.5% AD cases increase AD risk [CASP8-GGGAGA-AD-R1; OR 2.2, 95% CI (1.5185 to 3.1896), P = 3.1 × 10-5]. Cells transfected with a high-risk CASP8-GGGAGA-AD-R1 variant show increased toxicity and increased levels of poly(GR)n(GE)n(RE)n aggregates. Taken together, these data identify polyGR(+) aggregates as a frequent and unexpected type of brain pathology in AD and CASP8-GGGAGA-AD-R1 alleles as a relatively common AD risk factor. Taken together, these data support a model in which CASP8-GGGAGAEXP alleles combined with stress increase AD risk.
Keywords: Alzheimer’s disease; RAN proteins; microsatellite expansion mutations; protein aggregates; repeat associated non-AUG (RAN) translation.
Conflict of interest statement
Competing interests statement:T.E.G. is a cofounder and head of scientific advisory board for Andante Biologics. H.B.C. served as a consultant for Janssen Research and Development in 2021 and 2022. A.G. serves on the SAB of Genetech and Muna Therapeutics. B.T.H. serves on the SAB of Dewpoint and has an option for stock. He serves on a scientific advisory board or is a consultant for AbbVie, Alexion, Ambagon, Aprinoia Therapeutics, Arvinas, Avrobio, AstraZenica, Biogen, Bioinsights, BMS, Cure Alz Fund, Cell Signaling, Dewpoint, Latus, Novartis, Pfizer, Sanofi, Sofinnova, Vigil, Violet, Voyager, WaveBreak. T.E.G. owns stock and stock options in Andante Biologics. B.T.H. has an option for stock in Dewpoint. B.T.H. owns stock in Novartis. T.E.G. is an inventor on multiple patents and patent applications that relate to AD therapeutics. L.P.W.R. is an inventor on patents that are related to RAN translation. L.N. is an inventor on patents that are related to RAN translation.
Figures


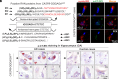


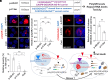
References
-
- Cacace R., Sleegers K., Van Broeckhoven C., Molecular genetics of early-onset Alzheimer’s disease revisited. Alzheimers Dement 12, 733–748 (2016). - PubMed
MeSH terms
Substances
Grants and funding
- R37 NS040389/NS/NINDS NIH HHS/United States
- K99AG065511 R00AG065511/HHS | NIH | National Institute on Aging (NIA)
- P30 AG066507/AG/NIA NIH HHS/United States
- R01NS122766/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- P30AG066506 P50AG047266/HHS | NIH | National Institute on Aging (NIA)
- U24 AG021886/AG/NIA NIH HHS/United States
- K99 AG065511/AG/NIA NIH HHS/United States
- R00 AG065511/AG/NIA NIH HHS/United States
- R01 NS122766/NS/NINDS NIH HHS/United States
- P50 AG047266/AG/NIA NIH HHS/United States
- W81XWH2210592/DOD | U.S. Army (USA)
- R37NS040389 RF1NS126536 RF1-NS098819/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- P30 AG066514/AG/NIA NIH HHS/United States
- RF1 NS126536/NS/NINDS NIH HHS/United States
- RF1 NS098819/NS/NINDS NIH HHS/United States
- NIH P30 AG0066507/HHS | NIH | National Institute on Aging (NIA)
- P30 AG066506/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

