Persistent postremission clonal hematopoiesis shapes the relapse trajectories of acute myeloid leukemia
- PMID: 39938015
- PMCID: PMC12008691
- DOI: 10.1182/bloodadvances.2024015149
Persistent postremission clonal hematopoiesis shapes the relapse trajectories of acute myeloid leukemia
Abstract
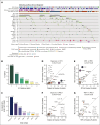
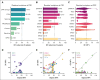
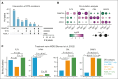
Mutations found in acute myeloid leukemia (AML) such as DNMT3A, TET2, and ASXL1 can be found in the peripheral blood of healthy adults, a phenomenon termed clonal hematopoiesis (CH). These mutations are thought to represent the earliest genetic events in the evolution of AML. Genomic studies on samples acquired at diagnosis, remission, and at relapse have demonstrated significant stability of CH mutations after induction chemotherapy. Meanwhile, later mutations in genes such as NPM1 and FLT3 have been shown to contract at remission, and in the case of FLT3 often are absent at relapse. We sought to understand how early CH mutations influence subsequent evolutionary trajectories throughout remission and relapse in response to induction chemotherapy. We assembled a retrospective cohort of patients diagnosed with de novo AML at our institution that underwent genomic sequencing at diagnosis, remission, and/or relapse (total N = 182 patients). FLT3 and NPM1 mutations were generally eliminated at complete remission but subsequently reemerged upon relapse, whereas DNMT3A, TET2, and ASXL1 mutations often persisted through remission. CH-related mutations exhibited distinct constellations of co-occurring genetic alterations, with NPM1 and FLT3 mutations enriched in DNMT3Amut AML, whereas CBL and SRSF2 mutations were enriched in TET2mut and ASXL1mut AML, respectively. In the case of NPM1 and FLT3 mutations, these differences vanished at the time of complete remission yet readily reemerged upon relapse, indicating the reproducible nature of these genetic interactions. Thus, CH-associated mutations that likely precede malignant transformation subsequently shape the evolutionary trajectories of AML through diagnosis, therapy, and relapse.
© 2025 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

