Linking obesity-associated genotype to child language development: the role of early-life neurology-related proteomics and brain myelination
- PMID: 39938231
- PMCID: PMC11868953
- DOI: 10.1016/j.ebiom.2025.105579
Linking obesity-associated genotype to child language development: the role of early-life neurology-related proteomics and brain myelination
Abstract
Background: The association between childhood obesity and language development may be confounded by socio-environmental factors and attributed to comorbid pathways.
Methods: In a longitudinal Singaporean mother-offspring cohort, we leveraged trans-ancestry polygenic predictions of body mass index (BMI) to interrogate the causal effects of early-life BMI on child language development and its effects on molecular and neuroimaging measures. Leveraging large genome-wide association studies, we examined whether the link between obesity and language development is causal or due to a shared genetic basis.
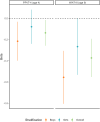
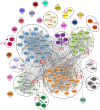
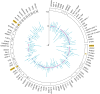
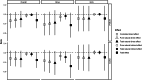
Findings: We found an inverse association between polygenic risk for obesity, which is less susceptible to confounding, and language ability assessed at age 9. Our findings suggested a shared genetic basis between obesity and language development rather than a causal effect of obesity on language development. Interrogating early-life mechanisms including neurology-related proteomics and language-related white matter microstructure, we found that EFNA4 and VWC2 expressions were associated with language ability as well as fractional anisotropy of language-related white matter tracts, suggesting a role in brain myelination. Additionally, the expression of the EPH-Ephrin signalling pathway in the hippocampus might contribute to language development. Polygenic risk for obesity was nominally associated with EFNA4 and VWC2 expression. However, we did not find support for mediating mechanisms via these proteins.
Interpretation: This study demonstrates the potential of examining early-life proteomics in conjunction with deep genotyping and phenotyping and provides biological insights into the shared genomic links between obesity and language development.
Funding: Singapore National Research Foundation and Agency for Science, Technology and Research.
Keywords: Language development; Neurology-related protein; Obesity; Polygenic risk score.
Copyright © 2025 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests KMG and SC are part of an academic consortium that has received research funding from Société Des Produits Nestlé S.A., and are co-inventors on patent filings by Nestlé S.A. outside the submitted work. KMG has received reimbursement for speaking at conferences sponsored by companies selling nutritional products. SC has received reimbursement from the Expert Group on Inositol in Basic and Clinical Research (EGOI; a not-for-profit academic organization) and Nestlé Nutrition Institute for speaking at conferences. The other authors declare no competing interests.
Figures

References
-
- Gibson J.L., Newbury D.F., Durkin K., Pickles A., Conti-Ramsden G., Toseeb U. Pathways from the early language and communication environment to literacy outcomes at the end of primary school; the roles of language development and social development. Oxf Rev Educ. 2021;47(2):260–283.
-
- Schoon I., Parsons S., Rush R., Law J. Children's language ability and psychosocial development: a 29-year follow-up study. Pediatrics. 2010;126(1):e73–e80. - PubMed
-
- Snowdon D.A., Greiner L.H., Markesbery W.R. Linguistic ability in early life and the neuropathology of Alzheimer's disease and cerebrovascular disease. Findings from the Nun Study. Ann N Y Acad Sci. 2000;903:34–38. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

