EXPERT expands prime editing efficiency and range of large fragment edits
- PMID: 39939583
- PMCID: PMC11822059
- DOI: 10.1038/s41467-025-56734-9
EXPERT expands prime editing efficiency and range of large fragment edits
Abstract
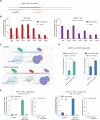
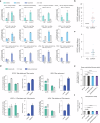
Prime editing systems (PEs) hold great promise in modern biotechnology. However, their editing range is limited as PEs can only modify the downstream sequences of the pegRNA nick. Here, we report the development of the extended prime editor system (EXPERT) to overcome this limitation by using an extended pegRNA (ext-pegRNA) with modified 3' extension, and an additional sgRNA (ups-sgRNA) targeting the upstream region of the ext-pegRNA. We demonstrate that EXPERT can efficiently perform editing on both sides of the ext-pegRNA nick, a task that is unattainable by canonical PEs. EXPERT exhibits prominent capacity in replacing sequences up to 88 base pairs and inserting sequences up to 100 base pairs within the upstream region of the ext-pegRNA nick. Compared to canonical PEs such as PE2, the utilization of the EXPERT strategy significantly enhances the editing efficiency for large fragment edits with an average improvement of 3.12-fold, up to 122.1 times higher. Safety wise, the use of ups-sgRNA does not increase the rates of undesirable insertions and deletions (indels), as the two nicks are on the same strand. Moreover, we do not observe increased off-target editing rates genome-wide. Our work introduces EXPERT as a PE tool with significant potential in life sciences.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: S.Z., J.R., Y.X., X.L., Y.S., R.H., X.X. (Xiaoning Xi), X.H., and S.X. have filed two patent applications related to this work through Huazhong Agricultural University (2023115813808, 2023115813795). The remaining authors declare no competing interests.
Figures



References
-
- Liang, R. et al. Prime editing using CRISPR-Cas12a and circular RNAs in human cells. Nat. Biotechnol.42,1867–1875 (2024). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

