Segment Anything for Microscopy
- PMID: 39939717
- PMCID: PMC11903314
- DOI: 10.1038/s41592-024-02580-4
Segment Anything for Microscopy
Erratum in
-
Author Correction: Segment Anything for Microscopy.Nat Methods. 2025 Jul;22(7):1603. doi: 10.1038/s41592-025-02745-9. Nat Methods. 2025. PMID: 40490528 Free PMC article. No abstract available.
Abstract
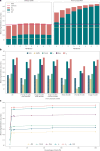
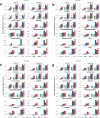
Accurate segmentation of objects in microscopy images remains a bottleneck for many researchers despite the number of tools developed for this purpose. Here, we present Segment Anything for Microscopy (μSAM), a tool for segmentation and tracking in multidimensional microscopy data. It is based on Segment Anything, a vision foundation model for image segmentation. We extend it by fine-tuning generalist models for light and electron microscopy that clearly improve segmentation quality for a wide range of imaging conditions. We also implement interactive and automatic segmentation in a napari plugin that can speed up diverse segmentation tasks and provides a unified solution for microscopy annotation across different microscopy modalities. Our work constitutes the application of vision foundation models in microscopy, laying the groundwork for solving image analysis tasks in this domain with a small set of powerful deep learning models.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures














References
-
- Schmidt, U., Weigert, M., Broaddus, C. & Myers, G. Cell detection with star-convex polygons. Lect. Notes Comput. Sci.11071, 265–273 (2018).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

