Learning produces an orthogonalized state machine in the hippocampus
- PMID: 39939774
- PMCID: PMC11964937
- DOI: 10.1038/s41586-024-08548-w
Learning produces an orthogonalized state machine in the hippocampus
Abstract
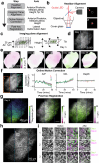
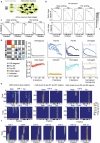
Cognitive maps confer animals with flexible intelligence by representing spatial, temporal and abstract relationships that can be used to shape thought, planning and behaviour. Cognitive maps have been observed in the hippocampus1, but their algorithmic form and learning mechanisms remain obscure. Here we used large-scale, longitudinal two-photon calcium imaging to record activity from thousands of neurons in the CA1 region of the hippocampus while mice learned to efficiently collect rewards from two subtly different linear tracks in virtual reality. Throughout learning, both animal behaviour and hippocampal neural activity progressed through multiple stages, gradually revealing improved task representation that mirrored improved behavioural efficiency. The learning process involved progressive decorrelations in initially similar hippocampal neural activity within and across tracks, ultimately resulting in orthogonalized representations resembling a state machine capturing the inherent structure of the task. This decorrelation process was driven by individual neurons acquiring task-state-specific responses (that is, 'state cells'). Although various standard artificial neural networks did not naturally capture these dynamics, the clone-structured causal graph, a hidden Markov model variant, uniquely reproduced both the final orthogonalized states and the learning trajectory seen in animals. The observed cellular and population dynamics constrain the mechanisms underlying cognitive map formation in the hippocampus, pointing to hidden state inference as a fundamental computational principle, with implications for both biological and artificial intelligence.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures













References
-
- O’Keefe, J. & Nadel, L. The Hippocampus as a Cognitive Map (1978).
-
- Tolman, E. C. Cognitive maps in rats and men. Psychol. Rev.55, 189–208 (1948). - PubMed
-
- O’Keefe, J. & Dostrovsky, J. The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res.34, 171–175 (1971). - PubMed
-
- O’Keefe, J. Place units in the hippocampus of the freely moving rat. Exp. Neurol.51, 78–109 (1976). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

