Uncovering two neutrophil-committed progenitors that immediately precede promyelocytes during human neutropoiesis
- PMID: 39939817
- PMCID: PMC11868371
- DOI: 10.1038/s41423-025-01259-w
Uncovering two neutrophil-committed progenitors that immediately precede promyelocytes during human neutropoiesis
Abstract
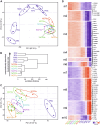
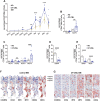
Technological advances have greatly improved our knowledge of myelopoiesis, for example, with the discovery of granulocyte‒monocyte‒dendritic cell (DC) progenitors (GMDPs), monocyte‒DC progenitors (MDPs), common DC progenitors (CDPs) and common monocyte progenitors (cMoPs) on the basis of flow cytometry approaches. Concomitantly, some progress has been made in characterizing the very early phases of human neutropoiesis with the description of novel CD66b+ progenitors, including eNePs, PMs w/o eNePs, ProNeus, and PreNeus. More recently, we identified four SSCloLin-CD66b-CD45dimCD34+/CD34dim/-CD64dimCD115- cells as the earliest precursors specifically committed to the neutrophil lineage present in human bone marrow (BM), which we called neutrophil-committed progenitors (NCPs, from NCP1s to NCP4s). In this study, we report the isolation and characterization of two new SSChiCD66b-CD64dimCD115-NCPs that, by phenotypic, transcriptomic, maturation and immunohistochemistry properties, as well as by flow cytometric side-scattered light (SSC), stand after NCP4s but precede promyelocytes during the neutropoiesis cascade. Similar to SSCloCD45RA+NCP2s/NCP3s and SSCloCD45RA-NCP1s/NCP4s, these cells exhibit phenotypic differences in CD45RA expression levels and, therefore, were named SSChiCD45RA+NCP5s and SSChiCD45RA-NCP6s. Moreover, NCP5s were more immature than NCP6s, as determined by cell differentiation and proliferative potential, as well as by transcriptomic and phenotypical features. Finally, by examining whether NCPs and all other CD66b+ neutrophil precursors are altered in representative hematological malignancies, we found that, in patients with chronic-phase chronic myeloid leukemia (CP-CML), but not with systemic mastocytosis (SM), there is an increased frequency of BM NCP4s, NCP6s, and all downstream CD45RA-negative neutrophil progenitors, suggesting their expansion in CML pathogenesis. Taken together, our data advance our knowledge of human neutropoiesis.
Keywords: Chronic myeloid leukemia; G-CSF; Neutrophils; Neutrophil progenitors; Neutropoiesis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Calzetti F, Finotti G, Cassatella MA. Current knowledge on the early stages of human neutropoiesis. Immunol Rev. 2023;314:111–24. - PubMed
-
- Calzetti F, Finotti G, Tamassia N, Bianchetto-Aguilera F, Castellucci M, Cane S, et al. CD66b(-)CD64(dim)CD115(-) cells in the human bone marrow represent neutrophil-committed progenitors. Nat Immunol. 2022;23:679–91. - PubMed
MeSH terms
Substances
Grants and funding
- IG-27613/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- PRIN20227YR8AW/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)
- PRIN2022W839FM/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

