Improvement of Mutant Galactose-1-Phosphate Uridylyltransferase (GALT) Activity by FDA-Approved Pharmacochaperones: A Preliminary Study
- PMID: 39940658
- PMCID: PMC11816840
- DOI: 10.3390/ijms26030888
Improvement of Mutant Galactose-1-Phosphate Uridylyltransferase (GALT) Activity by FDA-Approved Pharmacochaperones: A Preliminary Study
Abstract
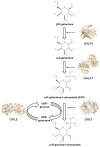
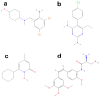
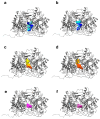
Classic galactosemia is a rare disease with long-term consequences that seriously affect the quality of life of patients. To date, various therapeutic approaches are being developed, but treatments that target the molecular defects in the mutant galactose-1-phosphate uridylyltransferase (GALT) gene are lacking. We conducted a computational search for putative pharmacochaperones by applying a drug repurposing strategy, and we found that one compound, already active as a pharmacochaperone in another pathology, doubled the enzymatic activity of the purified mutant enzyme in an in vitro test. Furthermore, an extensive computational search in a database of known active molecules found another compound able in its turn to improve in vitro enzymatic activity. Both compounds are predicted to interact with a cavity at the enzyme interface previously supposed to be an allosteric site for the GALT enzyme. In vitro tests confirmed also the reduced accumulation of galactose-1-phosphate (G1P) in fibroblasts of patients. Although these results must be considered preliminary, our findings pave the way for future research lines focused on the search for promising pharmacochaperones that can directly rescue the activity of the enzyme.
Keywords: classic galactosemia; drug repurposing; pharmacochaperones; protein mutations; rare diseases; virtual screening.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

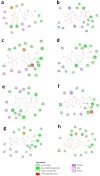
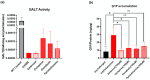
References
-
- Berry G.T. Classic Galactosemia and Clinical Variant Galactosemia. In: Adam M.P., Feldman J., Mirzaa G.M., Pagon R.A., Wallace S.E., Amemiya A., editors. GeneReviews® [Internet] University of Washington; Seattle, WA, USA: 1993–2024. [(accessed on 12 January 2025)]. Updated 11 March 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1518/ - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

