"Omics" and Postmortem Interval Estimation: A Systematic Review
- PMID: 39940802
- PMCID: PMC11817326
- DOI: 10.3390/ijms26031034
"Omics" and Postmortem Interval Estimation: A Systematic Review
Abstract
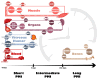
Postmortem interval (PMI) estimation is a challenge of utmost importance in forensic daily practice. Traditional methods face limitations in accuracy and reliability, particularly for advanced decomposition stages. Recent advances in "omics" sciences, providing a holistic view of postmortem biochemical changes, offer promising avenues for overcoming these challenges. This systematic review aims at investigating the role of mass-spectrometry-based "omics" approaches in PMI estimation to elucidate molecular mechanisms underlying predictable time-dependent biochemical alterations occurring after death. A systematic search was performed, adhering to PRISMA guidelines, through "free-text" protocols in the databases PubMed, SCOPUS and Web of Science. The inclusion criteria were as follows: experimental studies analyzing, as investigated samples, animal or human corpses in toto or in parts and estimating PMI through MS-based untargeted omics approaches, with full texts in the English language. Quality assessment was performed using STROBE and ARRIVE critical appraisal checklists. A total of 1152 papers were screened and 26 included. Seventeen papers adopted a proteomic approach (65.4%), nine focused on metabolomics (34.6%) and two on lipidomics (7.7%). Most papers (57.7%) focused on short PMIs (<7 days), the remaining papers explored medium (7-120 days) (30.77%) and long PMIs (>120 days) (15.4%). Muscle tissue was the most frequently analyzed substrate (34.6% of papers), followed by liver (19.2%), bones (15.4%), cardiac blood and leaking fluids (11.5%), lung, kidney and serum (7.7%), and spleen, vitreous humor and heart (3.8%). Predictable time-dependent degradation patterns of macromolecules in different biological substrates have been discussed, with special attention to molecular insights into postmortem biochemical changes.
Keywords: forensic; lipidomics; mass-spectrometry; metabolomics; omics; postmortem interval; proteomics; time-since-death.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Brockbals L., Garrett-Rickman S., Fu S., Ueland M., McNevin D., Padula M.P. Estimating the Time of Human Decomposition Based on Skeletal Muscle Biopsy Samples Utilizing an Untargeted LC–MS/MS-Based Proteomics Approach. Anal. Bioanal. Chem. 2023;415:5487–5498. doi: 10.1007/s00216-023-04822-4. - DOI - PMC - PubMed
-
- Nolan A.N., Mead R.J., Maker G., Bringans S., Chapman B., Speers S.J. Examination of the Temporal Variation of Peptide Content in Decomposition Fluid under Controlled Conditions Using Pigs as Human Substitutes. Forensic Sci. Int. 2019;298:161–168. doi: 10.1016/j.forsciint.2019.02.048. - DOI - PubMed
-
- Burkhard M. Handbook of Forensic Medicine. 1st ed. Wiley Blackwell; Hoboken, NJ, USA: 2013. pp. 75–133.
-
- Dettmeyer R.B., Verhoff M.A., Schütz H.F. Forensic Medicine. Fundamentals and Perspectives. Springer; Berlin/Heidelberg, Germany: 2014. pp. 33–55.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources