Improved Recombinant Adeno-Associated Viral Vector Production via Molecular Evolution of the Viral Rep Protein
- PMID: 39941089
- PMCID: PMC11818820
- DOI: 10.3390/ijms26031319
Improved Recombinant Adeno-Associated Viral Vector Production via Molecular Evolution of the Viral Rep Protein
Abstract
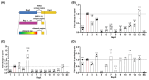
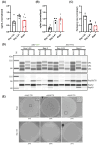
In the dynamic field of gene therapy, recombinant adeno-associated viruses (rAAVs) have become leading viral vectors due to their safety, long-term expression, and wide-ranging cell and tissue tropism. With numerous FDA approvals and commercial products underscoring their potential, there is a critical need for efficient production processes to achieve high vector titers and quality. A major challenge in rAAV production is the efficient packaging of the genome into the viral capsid, with empty or partially filled capsids often representing over 90% of the produced material. To tackle this issue, we engineered the replication and packaging proteins of an AAV (Rep) to boost their functionality and improve vector titers. We subjected a complex Rep library derived from the AAV serotypes 1-13 to directed evolution in an AAV producer cell line. After each round of selection, single clones were analyzed, showing enrichment of specific hybrid Rep domains. Comparative analysis of these selected clones revealed considerable differences in their ability to package AAV2-based viral genomes, with hybrid Rep proteins achieving up to a 2.5-fold increase in packaging efficiency compared to their parental counterparts. These results suggest that optimizing rep gene variants through directed evolution is an effective strategy to enhance rAAV production efficiency.
Keywords: AAV; directed evolution; protein engineering; rAAV production; replication proteins.
Conflict of interest statement
Authors Thomas Steininger, Veronika Öttl, Cornelius Frank, Johannes Fritsch, Evelyn Hirschauer, Fabian Konkel, Sabine Linder, Stefan Seeber, Julia Fakhiri, Philip Ohland, Markus Haindl, Gregor Pechmann, Marie-Sofie Dürr, Stefan Klostermann, Anna Sandmeir, Luisa D. Hilgenfeld, Florian Semmelmann are employed by Roche Diagnostics GmbH, Linda E. Franken, Philip Ohland, Miriam Lopez Ferreiro, Philippe Ringler, Matthias E. Lauer, Denis Phichith are employed by Hoffmann-La Roche Ltd., Mustafa N. Yazicioglu is employed by Roche Holding AG. A patent application related to the research has been submitted and is currently awaiting approval. The authors declare no competing interests.
Figures



References
-
- Williams C. At-Cost AAV: From Production Cost to Patient Dose. [(accessed on 20 January 2025)]. Available online: https://desciappliedresearch.com/2024/01/at-cost-aav/
-
- McIntosh N.L., Berguig G.Y., Karim O.A., Cortesio C.L., De Angelis R., Khan A.A., Gold D., Maga J.A., Bhat V.S. Comprehensive characterization and quantification of adeno associated vectors by size exclusion chromatography and multi angle light scattering. Sci. Rep. 2021;11:3012. doi: 10.1038/s41598-021-82599-1. - DOI - PMC - PubMed
-
- Determination of Full, Partial and Empty Capsid Ratios for Adeno-Associated Virus (AAV) Analysis. [(accessed on 20 January 2025)]. Available online: https://sciex.com/tech-notes/ce/determination-of-full--partial-and-empty....
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

