Association of Inflammatory Profile During Ex Vivo Lung Perfusion With High-Grade Primary Graft Dysfunction: A Systematic Review and Meta-Analysis
- PMID: 39944219
- PMCID: PMC11815944
- DOI: 10.3389/ti.2025.13794
Association of Inflammatory Profile During Ex Vivo Lung Perfusion With High-Grade Primary Graft Dysfunction: A Systematic Review and Meta-Analysis
Abstract
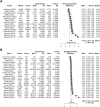
PGD3 is the manifestation of ischemia-reperfusion injury which results from inflammation and cell death and is associated with poor outcome. This systematic-review and meta-analysis of non-randomized controlled trials on patients undergoing Ltx with reconditioned lungs via EVLP, aims to assess the association between the levels of proinflammatory biomarkers during EVLP and PGD3 development within the firsts 72 h post-Ltx. Biomarkers were categorized by timing (1-hour, T0 and 4-hours, Tend from EVLPstart) and by their biological function (adhesion molecules, chemokines, cytokines, damage-associated-molecular-patterns, growth-factors, metabolites). We employed a four-level mixed-effects model with categorical predictors for biomarker groups to identify differences between patients with PGD3 and others. The single study and individual measurements were considered random intercepts. We included 8 studies (610 measurements at T0 and 884 at Tend). The pooled effect was 0.74 (p = 0.021) at T0, and 0.90 (p = 0.0015) at Tend. The four-level model indicated a large pooled correlation between developing PGD3 at 72 h post-Ltx and inflammatory biomarkers values, r = 0.62 (p = 0.009). Chemokine group showed the strongest association with the outcome (z-value = 1.26, p = 0.042). Pooled panels of inflammation markers, particularly chemokines, measured at T0 or at Tend, are associated with the development of PGD3 within the first 72 h after Ltx.
Systematic review registration: https://osf.io/gkxzh/.
Keywords: biomarkers; ex vivo lung perfusion; inflammation; ischemia-reperfusion injury; primary graft dysfunction.
Copyright © 2025 Costamagna, Balzani, Marro, Simonato, Burello, Rinaldi, Brazzi, Boffini and Fanelli.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

